Abstract
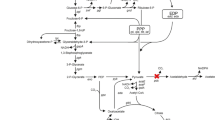
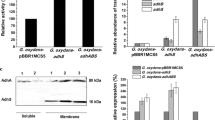
Acetic acid bacteria such as Gluconobacter oxydans are used in several biotechnological processes due to their ability to perform rapid incomplete regio- and stereo-selective oxidations of a great variety of carbohydrates, alcohols, and related compounds by their membrane-bound dehydrogenases. In order to understand the growth physiology of industrial strains such as G. oxydans ATCC 621H that has high substrate oxidation rates but poor growth yields, we compared its genome sequence to the genome sequence of strain DSM 3504 that reaches an almost three times higher optical density. Although the genome sequences are very similar, DSM 3504 has additional copies of genes that are absent from ATCC 621H. Most importantly, strain DSM 3504 contains an additional type II NADH dehydrogenase (ndh) gene and an additional triosephosphate isomerase (tpi) gene. We deleted these additional paralogs from DSM 3504, overexpressed NADH dehydrogenase in ATCC 621H, and monitored biomass and the concentration of the representative cell components as well as O2 and CO2 transfer rates in growth experiments on mannitol. The data revealed a clear competition of membrane-bound dehydrogenases and NADH dehydrogenase for channeling electrons in the electron transport chain of Gluconobacter and an important role of the additional NADH dehydrogenase for increased growth yields. The less active the NADH dehydrogenase is, the more active is the membrane-bound polyol dehydrogenase. These results were confirmed by introducing additional ndh genes via plasmid pAJ78 in strain ATCC 621H, which leads to a marked increase of the growth rate.




Similar content being viewed by others
References
Anderlei T, Zang W, Papaspyrou E, Büchs J (2004) Online respiration activity measurement (OTR, CTR, RQ) in shake flasks. Biochem Eng J 17:187–194
Cava F, Zafra O, Magalon A, Blasco F, Berenguer J (2004) A new type of NADH dehydrogenase specific for nitrate respiration in the extreme thermophile Thermus thermophilus. J Biol Chem 279(44):45369–45378
De Ley J, Swings J, Gossele F (1984) The genus Gluconobacter. In: Krieg N, Holt J (eds) Bergey’s manual of systematic bacteriology. Williams & Wilkins Co., Baltimore, pp 267–278
De Muynck C, Pereira CS, Naessens M, Parmentier S, Soetaert W, Vandamme EJ (2007) The genus Gluconobacter oxydans: comprehensive overview of biochemistry and biotechnological applications. Crit Rev Biotechnol 27(3):147–171
Delcher AL, Harmon D, Kasif S, White O, Salzberg SL (1999) Improved microbial gene identification with GLIMMER. Nucleic Acids Res 27(23):4636–4641
Deppenmeier U, Ehrenreich A (2009) Physiology of acetic acid bacteria in light of the genome sequence of Gluconobacter oxydans. J Mol Microbiol Biotechnol 16(1–2):69–80
Deppenmeier U, Hoffmeister M, Prust C (2002) Biochemistry and biotechnological applications of Gluconobacter strains. Appl Microbiol Biotechnol 60(3):233–242
Elfari M, Ha S-W, Bremus C, Merfort M, Khodaverdi V, Herrmann U, Sahm H, Görisch H (2005) A Gluconobacter oxydans mutant converting glucose almost quantitatively to 5-keto-D-gluconic acid. Appl Microbiol Biotechnol 66(6):668–674
Friedrich T, Scheide D (2000) The respiratory complex I of bacteria, archaea and eukarya and its module common with membrane-bound multisubunit hydrogenases. FEBS Lett 479(1–2):1–5
Gillis M, De Ley J (1980) Intra- and intergenic similarities of the ribosomal ribonucleic acid cistrons of Acetobacter and Gluconobacter. Int J Sys Bacteriol 30:7–27
Gupta A, Singh VK, Qazi GN, Kumar A (2001) Gluconobacter oxydans: its biotechnological applications. J Mol Microbiol Biotechnol 3(3):445–456
Hall PE, Anderson SM, Johnston DM, Cannon RE (1992) Transformation of Acetobacter xylinum with plasmid DNA by electroporation. Plasmid 28(3):194–200
Hanahan D (1983) Studies on transformation of Escherichia coli with plasmids. J Mol Biol 166(4):557–580
Huber R, Palmen TG, Ryk N, Hillmer AK, Luft K, Kensy F, Büchs J (2010) Replication methods and tools in high-throughput cultivation processes—recognizing potential variations of growth and product formation by on-line monitoring. BMC Biotechnol 10:22
Jaworowski A, Campbell HD, Poulis MI, Young IG (1981) Genetic identification and purification of the respiratory NADH dehydrogenase of Escherichia coli. Biochemistry 20(7):2041–2047
Kallnik V, Meyer M, Deppenmeier U, Schweiger P (2010) Construction of expression vectors for protein production in Gluconobacter oxydans. J Biotechnol 150(4):460–465
Kensy F, Zang E, Faulhammer C, Tan RK, Büchs J (2009) Validation of a high-throughput fermentation system based on online monitoring of biomass and fluorescence in continuously shaken microtiter plates. Microb Cell Fact 8:31
Knowles JR (1991) Enzyme catalysis: not different, just better. Nature 350(6314):121–124
Kostner D, Peters B, Mientus M, Liebl W, Ehrenreich A (2013) Importance of codB for new codA-based markerless gene deletion in Gluconobacter strains. Appl Microbiol Biotechnol 97(18):8341–8349
Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM, Peterson KM (1995) Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166(1):175–176
Lagesen K, Hallin P, Rodland EA, Staerfeldt HH, Rognes T, Ussery DW (2007) RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res 35(9):3100–3108
Levering PR, Weenk G, Olijve W, Dijkhuizen L, Harder W (1988) Regulation of gluconate and ketogluconate production in Gluconobacter oxydans ATCC 621-H. Arch Microbiol 149:534–539
Lowe TM, Eddy SR (1997) tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25(5):955–964
Markowitz VM, Mavromatis K, Ivanova NN, Chen IM, Chu K, Kyrpides NC (2009) IMG ER: a system for microbial genome annotation expert review and curation. Bioinformatics 25(17):2271–2278
Matsushita K, Shinagawa E, Ameyama M (1982) D-Gluconate dehydrogenase from bacteria, 2-keto-D-gluconate-yielding, membrane-bound. Methods Enzymol 89 Pt D:187–193
Matsushita K, Toyama H, Adachi O (1994) Respiratory chains and bioenergetics of acetic acid bacteria. Adv Microb Physiol 36:247–301
Meyer M, Schweiger P, Deppenmeier U (2013) Effects of membrane-bound glucose dehydrogenase overproduction on the respiratory chain of Gluconobacter oxydans. Appl Microbiol Biotechnol 97(8):3457–3466
Mostafa HE, Heller KJ, Geis A (2002) Cloning of Escherichia coli lacZ and lacY genes and their expression in Gluconobacter oxydans and Acetobacter liquefaciens. Appl Environ Microbiol 68(5):2619–2623
Motizuki K, Takarazuka I, Kanzaki T (1966) Method for producing 2-keto-L-gulonic acid
Nantapong N, Kugimiya Y, Toyama H, Adachi O, Matsushita K (2004) Effect of NADH dehydrogenase-disruption and over-expression on respiration-related metabolism in Corynebacterium glutamicum KY9714. Appl Microbiol Biotechnol 66(2):187–193
Overbeek R, Larsen N, Walunas T, D'Souza M, Pusch G, Selkov E, Liolios K, Joukov V, Kaznadzey D, Anderson I, Bhattacharyya A, Burd H, Gardner W, Hanke P, Kapatral V, Mikhailova N, Vasieva O, Osterman A, Vonstein V, Fonstein M, Ivanova N, Kyrpides N (2003) The ERGO genome analysis and discovery system. Nucleic Acids Res 31(1):164–171
Peters B, Junker A, Brauer K, Mühlthaler B, Kostner D, Mientus M, Liebl W, Ehrenreich A (2013a) Deletion of pyruvate decarboxylase by a new method for efficient markerless gene deletions in Gluconobacter oxydans. Appl Microbiol Biotechnol 97(6):2521–2530
Peters B, Mientus M, Kostner D, Junker A, Liebl W, Ehrenreich A (2013b) Characterization of membrane-bound dehydrogenases from Gluconobacter oxydans 621H via whole-cell activity assays using multideletion strains. Appl Microbiol Biotechnol 97(14):6397–6412
Prust C, Hoffmeister M, Liesegang H, Wiezer A, Fricke WF, Ehrenreich A, Gottschalk G, Deppenmeier U (2005) Complete genome sequence of the acetic acid bacterium Gluconobacter oxydans. Nat Biotechnol 23(2):195–200
Richhardt J, Bringer S, Bott M (2012) Mutational analysis of the pentose phosphate and Entner-Doudoroff pathways in Gluconobacter oxydans reveals improved growth of a Δedd Δeda mutant on mannitol. Appl Environ Microbiol 78(19):6975–6986
Richhardt J, Luchterhand B, Bringer S, Büchs J, Bott M (2013) Evidence for a key role of cytochrome bo 3 oxidase in respiratory energy metabolism of Gluconobacter oxydans. J Bacteriol 195(18):4210–4220
Rose IA, Fung WJ, Warms JV (1990) Proton diffusion in the active site of triosephosphate isomerase. Biochemistry 29(18):4312–4317
Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B (2000) Artemis: sequence visualization and annotation. Bioinformatics 16(10):944–945
Sambrook J, Fritsch E, Maniatis T (1989) Molecular cloning a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Samorski M, Muller-Newen G, Büchs J (2005) Quasi-continuous combined scattered light and fluorescence measurements: a novel measurement technique for shaken microtiter plates. Biotechnol Bioeng 92(1):61–68
Schweiger P, Volland S, Deppenmeier U (2007) Overproduction and characterization of two distinct aldehyde-oxidizing enzymes from Gluconobacter oxydans 621H. J Mol Microbiol Biotechnol 13(1–3):147–155
Staden R, Beal KF, Bonfield JK (2000) The Staden package, 1998. Methods Mol Biol 132:115–130
Voss J, Ehrenreich A, Liebl W (2010) Characterization and inactivation of the membrane-bound polyol dehydrogenase in Gluconobacter oxydans DSM 7145 reveals a role in meso-erythritol oxidation. Microbiology (Reading, England)
Wach A (1996) PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. Yeast 12(3):259–265
Acknowledgments
We thank the Bundesministerium für Bildung und Forschung (BMBF) for funding this work in the framework of the GenoMik-Transfer initiative (FKZ: 0315632C).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 1255 kb)
Rights and permissions
About this article
Cite this article
Kostner, D., Luchterhand, B., Junker, A. et al. The consequence of an additional NADH dehydrogenase paralog on the growth of Gluconobacter oxydans DSM3504. Appl Microbiol Biotechnol 99, 375–386 (2015). https://doi.org/10.1007/s00253-014-6069-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-014-6069-9