Abstract
Interest in producing biofuels from renewable sources has escalated due to energy and environmental concerns. Recently, the production of higher chain alcohols from 2-keto acid pathways has shown significant progress. In this paper, we demonstrate a mutagenesis approach in developing a strain of Escherichia coli for the production of 3-methyl-1-butanol by leveraging selective pressure toward l-leucine biosynthesis and screening for increased alcohol production. Random mutagenesis and selection with 4-aza-d,l-leucine, a structural analogue to l-leucine, resulted in the development of a new strain of E. coli able to produce 4.4 g/L of 3-methyl-1-butanol. Investigation of the host’s sensitivity to 3-methyl-1-butanol directed development of a two-phase fermentation process in which titers reached 9.5 g/L of 3-methyl-1-butanol with a yield of 0.11 g/g glucose after 60 h.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Growing concerns with the diminishing reserves, supply security, and environmental impact of petroleum-based liquid fuels have intensified efforts to meet these challenges (Connor and Liao 2009; Dinneen 2008; Stephanopoulos 2007). Biofuels produced from renewable sources have the potential to mitigate some of these issues. The production of ethanol, in particular, has steadily risen to 9.2 billion gallons in 2008 in the USA (Seiferlein 2009), with new research directed toward using lignocellulosic feedstocks as a raw material as this would greatly increase the land yield for production and reduce the dependence of fuel production on food crops. Bioethanol production is an attractive option because of the high productivity and yield during fermentation. Ethanol, however, is limited by its low energy density (∼70% of gasoline) and hygroscopicity, making it incompatible with the current infrastructure. In hopes of addressing some of the shortcomings of ethanol, potential alternative fuels such as higher chain alcohols produced through the keto acid pathways (Atsumi et al. 2008a, b; Cann and Liao 2008; Connor and Liao 2008; Shen and Liao 2008; Zhang et al. 2008) or through engineered and natural fermentative pathways (Atsumi et al. 2008b; Lee et al. 2008) have been investigated. Higher chain alcohols (C3–C5), in particular, are attractive fuel alternatives due to their increased energy density and low hygroscopicity.
Previously, a metabolic engineering approach was applied to Escherichia coli for the production of the potential biofuel 3-methyl-1-butanol (3MB; Connor and Liao 2008). Metabolic engineering is the practice by which the development of biological processes is optimized through genetic and regulatory manipulations. This approach has been used for many years in the design of microorganisms for the production of many valuable metabolites and commodity chemicals and, more recently, biofuels (Atsumi et al. 2008a, b; Atsumi and Liao 2008; Kim et al. 2007; Lee et al. 2009; Withers et al. 2007; Yan and Liao 2009; Zhang et al. 2008). The use of rational design in the development of a biocatalyst, although widely successful, suffers from a few shortcomings, the most significant being that the metabolic pathway for the production of the target must be elucidated. For effective optimization, details on the genes and enzymes responsible for the synthesis of the target compound, as well as undesired side products, must be known. Cellular metabolism is very complex and often highly interdependent, and it is therefore difficult to account for every detail when designing a strain in this manner. One way to address this concern is to create a random library of strain variants and use selective pressure to discover strains with the desired phenotype. The use of selective pressure can allow for investigation on the order of 109–1012 strains, which would not be possible otherwise. This procedure can also be used iteratively to develop strains with increasingly desirable phenotypes. The difficulty in using this strategy is the development of a selection as this is not possible for every process. However, because the production of 3MB uses the 2-keto acid pathways, the development of a new strain can be greatly aided by the vast knowledge and technology base in amino acid production.
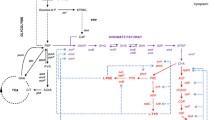
A powerful tool used in the development of amino acid production strains is random mutagenesis and selection with an amino acid analogue (Ikeda 2003). Chemical mutagens such as N-methyl-N′-nitro-N-nitrosoguanidine (NTG) or ethyl methanesulfonate are often used to create genome-wide mutations, with NTG being the more popular choice due to its superior mutation rate. Selection pressure in this case is provided by the toxicity of amino acid analogues. This toxicity could be due to several effects, including interference in the regulation of amino acid biosynthesis and incorporation into polypeptides. Among the means with which a cell can adapt to survive the challenge of the analogue is the ability to produce the natural amino acid in high enough quantities to outcompete the analogue for incorporation into growing polypeptides. Because this is not the only reason a cell can gain resistance to an analogue, variant strains which show an increased resistance to the amino acid analogue must then be screened and verified for the desired phenotype before being subjected to additional rounds of mutagenesis, if so desired. The advantage of this strategy is that the nearly infinite possibility of created phenotypes can effectively be studied because a selective strategy is being used. This strategy was applied to the production of 3MB which exploits the l-leucine biosynthesis pathway (Fig. 1). Here, we demonstrate the development of a strain of E. coli for the production of 3MB by subjecting a wild-type strain to mutagenesis by NTG and challenging these cells with 4-aza-d,l-leucine (AZL), a structural analogue to natural l-leucine (Table 1).
Materials and methods
Bacteria strains, media, and growth conditions
JCL16 (rrnB T14 ΔlacZ WJ16 hsdR514 ΔaraBAD AH33 ΔrhaBAD LD78/F′ [traD36, proAB+, lacIq ZΔM15]) was used as wild type (WT; Table 2).
To compare JCL16, AL1, and AL2 (Table 2), strains were grown in M9 medium (6 g Na2HPO4, 3 g KH2PO4, 1 g NH4Cl, 0.5 g NaCl, 1 mM MgSO4, 1 mM CaCl2, 10 mg vitamin B1 per liter of water) containing 40 g/L of glucose, 5 g/L of yeast extract, and 1000X A5 trace metals mix (2.86 g H3BO3, 1.81 g MnCl2⋅4H2O, 0.222 g ZnSO4⋅7H2O, 0.39 g Na2MoO4⋅2H2O, 0.079 g CuSO4⋅5H2O, and 49.4 mg Co(NO3)2⋅6H2O per liter water). From a 3 mL Luria–Bertani (LB) overnight, cultures were inoculated 1% by volume (v/v) into 20 mL of fresh medium in 250-mL screw cap flasks and grown at 37°C in a rotary shaker until the OD600 reached 0.4–0.8. The culture was then induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) and grown at 30°C for the remainder of the experiment. Antibiotics were added to all cultures (ampicillin 100 μg/mL, chloroamphenicol 35 μg/mL, and kanamycin 50 μg/mL).
For production experiments with AL2, 3 mL LB overnight cultures were used to inoculate 10 mL (1%, v/v) of M9 medium containing 85 g/L of glucose, 10 g/L of yeast extract, and 1000X A5 trace metals mix and were inoculated 1% from a 3 mL LB overnight into a 250-mL screw cap flask. Ten milliliters of oleyl alcohol was added to flasks prior to incubation for extraction experiments. Cultures were grown at 37°C in a rotary shaker until the OD600 reached 0.4–0.8 and were then induced with 1 mM IPTG and incubated at 30°C in a rotary shaker. For extraction experiments, equal volumes of the oil and aqueous layers were always sampled to maintain the volume ratio. Antibiotics were added to all cultures (ampicillin 100 μg/mL, chloroamphenicol 35 μg/mL, and kanamycin 50 μg/mL).
To determine the viable cell count of AL2 after exposure to 3MB, a 3 mL LB overnight was used to inoculate 100 mL (1%, v/v) of M9 medium containing 85 g/L of glucose, 10 g/L of yeast extract, and 1000X A5 trace metals mix into two 500-mL baffled shake flasks (50 mL each). Cultures were grown at 30°C in a rotary shaker until the OD600 was 0.4–0.8 and then split into 15-mL screw cap tubes containing 4 mL each. 3MB was then added to the tubes in various concentrations and incubated at 30°C in a rotary shaker. Each 3MB concentration was tested in duplicate tubes. Samples were taken at 8 and 24 h in which cultures were diluted in triplicate between 10−5 and 10−6 and plated on LB agar plates and incubated overnight at 37°C. The viable cell count was measured by counting the colony forming units (CFU) on each plate and normalizing it to the control (0 g/L 3MB).
Reagents
NTG and AZL were purchased from Sigma-Aldrich (St. Louis, MO, USA). Alcohol standards were purchased from Fisher Scientific (Pittsburgh, PA, USA) and Sigma-Aldrich. Oleyl alcohol was purchased from Sigma-Aldrich. High-performance liquid chromatography (HPLC) grade heptane was purchased from Fisher Scientific.
Mutagenesis, selection, and screening of mutant libraries
The protocol for NTG mutagenesis was adapted from the method of Miller (1972). From 3 mL LB overnight cultures of the parental strains (JCL16, AL1), cultures were inoculated 1% (v/v) into 5 mL of fresh LB medium and grown at 37°C in a rotary shaker until the OD600 reached 0.4–0.6. Cells were spun down at room temperature and washed with an equal volume 0.1 M Na citrate (pH 5.5) twice before resuspending the cells in half the original volume of 0.1 M Na citrate. NTG was added to a final concentration of 50 μg/mL from a 1 mg/mL stock in 0.1 M Na citrate and incubated at 37°C for 15 min. For the control experiment, an equal volume of 0.1 M Na citrate was added instead of NTG and incubated at 37°C for 15 min. After incubation, cells were washed twice with the original volume of 0.1 M phosphate buffer (pH 7.1). The cells were then resuspended in 5 mL of LB and incubated at 37°C overnight for outgrowth. A small amount of cells were plated for both the NTG-treated tube and the control tube on LB plates to determine the kill count for the experiment before outgrowth (Table 3). After outgrowth overnight, cultures were spun down and resuspended in 0.1 M phosphate buffer (pH 7.1) and plated on M9 plates containing 10 g/L of glucose, 1000X A5 trace metals, and 2 g/L (JCL16 mutagenesis) or 8 g/L (AL1 mutagenesis) of AZL and incubated at 37°C for 2–3 days.
Mutant colonies resistant to AZL were then re-streaked on identical plates to assure a stable phenotype. Colonies with this stable phenotype were then picked and used to inoculate 400 μL of M9 medium containing 10 g/L of glucose, 1000X A5 trace metals, and 1 g/L (JCL16 mutagenesis) or 6 g/L (AL1 mutagenesis) of AZL in 96-deep-well plates (Corning Inc., Corning, NY, USA). Plates were quantified for growth after 24 h of incubation at 37°C in a rotary shaker.
Mutant strains showing increased growth were then transformed with pIAA12 (Table 2) to screen for 3MB production. 3 mL LB overnight cultures of transformant strains were used to inoculate 10 mL (1%, v/v) of M9 medium containing 10 g/L of glucose, 5 g/L of yeast extract, and 1000X A5 trace metals in a 125-mL screw cap flask and incubated at 37°C in a rotary shaker until the OD600 was 0.4–0.8. Cultures were then induced with 1 mM IPTG and incubated at 30°C in a rotary shaker and sampled at 24 h for growth and alcohol production. Ampicillin (100 μg/mL) was added to all cultures.
Variant strains showing increased production of 3MB relative to the parental strain were then transformed with pIAA11, pIAA13, and pIAA16 (Table 2). Three milliliters LB overnights of these strains were used to inoculate 20 mL (1%, v/v) of M9 medium containing 30 g/L of glucose, 5 g/L of yeast extract, and 1000X A5 trace metals in a 250-mL screw cap flask and incubated at 37°C in a rotary shaker until the OD600 reached 0.4–0.8. Cultures were induced with 1 mM IPTG and incubated at 30°C in a rotary shaker and sampled at 24 and 48 h for growth and alcohol production. Antibiotics were added to all cultures (ampicillin 100 μg/mL, chloroamphenicol 35 μg/mL, and kanamycin 50 μg/mL).
Detection of metabolites
The produced alcohol compounds were quantified by a gas chromatograph (GC) equipped with a flame ionization detector. The system consisted of model 5890A GC (Hewlett Packard, Avondale, PA, USA) and a model 7673A automatic injector, sampler, and controller (Hewlett Packard). The separation of alcohol compounds was carried out by a DB-WAX capillary column (60 m, 0.32-mm i.d., 0.50-μm film thickness) purchased from Agilent Technologies (Santa Clara, CA, USA). GC oven temperature started at 40°C and was raised with a gradient of 5°C/min until 80°C and held for 2 min. It was then raised with a gradient of 45°C/min until 125°C and held for 2 min. The oven temperature was then raised at a rate of 60°C/min until 235°C and held for 5 min. Helium was used as the carrier gas with 25-psi inlet pressure. The injector and detector were maintained at 225°C. For aqueous samples, 0.5 μL of culture broth supernatant was injected in split injection mode. For oleyl alcohol samples, samples were diluted with HPLC grade heptane in a 1:1 ratio to reduce the viscosity and increase the accuracy of injection. Glucose was quantified using a YSI 2700 SELECT Biochemistry Analyzer (YSI Life Sciences, Yellow Springs, OH, USA).
Results
Mutagenesis with NTG and selection with AZL
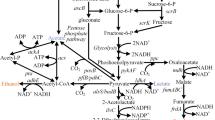
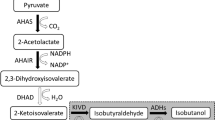
A schematic for the mutagenesis and selection/screening process is diagrammed in Fig. 2. In order to develop a new strain for the hyperproduction of 3MB, E. coli strain JCL16 (Table 2) was treated with NTG (see “Materials and methods”) and plated on minimal medium plates containing glucose (10 g/L) and AZL (2–8 g/L). Mutants displaying resistance to AZL were re-streaked on the same minimal medium plates to validate the phenotype. Resistant strains were then screened for growth in liquid minimal medium containing AZL. Growth was quantified after 24 h, and strains able to grow to a higher cell density were selected for transformation with kivd (Lactococcus lactis) and ADH2 (Saccharomyces cerevisiae) expressed from PL-lacO1 (pIAA12; Table 2). The production of 3MB from each of these strains was quantified after 24 h, and the strains able to produce the highest amount of 3MB were chosen for a final round of screening in which each of these strains was tested for 3MB production after overexpression of all the genes involved in the 3MB production pathway, including alsS (Bacillus subtilis), ilvCD (E. coli), leuA(G462D) (E. coli; Gusyatiner et al. 2002), and leuBCD (E. coli). These genes were all expressed from PL-lacO1 (pIAA11, pIAA16) along with kivd and ADH2 (pIAA13; Table 2). 3MB was quantified after 24 and 48 h, and the mutant that produced the greatest amount of 3MB was chosen for further analysis.
Diagram for mutagenesis/screening scheme. Cells are exposed to N-methyl-N′-nitro-N-nitrosoguanidine and selected for resistance to 4-aza-d,l-leucine on minimal media plates. Resistant colonies are screened for growth in liquid medium in 96-well plates (3), and assayed for 3MB production with kivd and ADH2 (5). Mutants showing an increase in production of 3MB are then assayed for production of 3MB with the entire pathway of genes (7). The highest producing mutant from this group is chosen as the parent for the next round of mutagenesis
Mutagenesis results
After the first round of mutagenesis and selection with 2 g/L of AZL, a total of 128 resistant colonies were isolated and re-streaked. Of these 128 variant strains, 21 were able to grow to at least OD600 = 0.4 after 24 h in minimal medium in the presence of 1 g/L of AZL as cells showed an increased sensitivity to the analogue in liquid medium compared to solid medium. These mutants were transformed with kivd and ADH2 (pIAA12) and assayed for 3MB production. Five mutants displayed a significant increase in 3MB production relative to parental strain (120 mg/L), and after transformation with the entire 3MB production pathway in these mutants, strain JCL16 NTG M8, named AL1, was able to produce nearly double the 3MB (0.75 g/L) as that of the WT (0.43 g/L). AL1 was then selected as the parent for the next round of mutagenesis. AL1 was treated with NTG as before and selection was carried out in the presence of 8 g/L of AZL. After the second round of mutagenesis, 64 colonies resistant to 8 g/L of AZL on solid medium were screened for growth in liquid media with 6 g/L of AZL, out of which 12 showed superior growth (OD600 > 0.25). Six mutants outperformed the parent upon transformation of kivd and ADH2 and were chosen for transformation with the entire 3MB pathway. Strain JCL16 NTG M8-C2, designated AL2, was able to produce a significantly higher amount (1.5 g/L) of 3MB of that AL1 after 24 h. AL2 was the only mutant from the second round to show an increase in 3MB production under this condition.
AL2 was then chosen as the parent for further mutagenesis. However, several attempts to generate a better host for 3MB production were unsuccessful. Strain AL2 was then compared with AL1 and JCL16 for growth, glucose consumption, and alcohol production (Fig. 3). Growth for each strain was similar, as was isobutanol production. AL1 was able to produce twice (1.1 g/L) the 3MB after 72 h as that of JCL16 (0.45 g/L) at a significantly higher yield (0.08 to 0.04 g/g after 36 h). AL2, however, showed a significant increase in glucose consumption and 3MB production relative to both strains. AL2 was able to produce 2.8 g/L of 3MB after 36 h at a yield of 0.11 g/g, which is significantly higher than either parental strains in terms of final product titer and yield. After 36 h, however, glucose consumption and 3MB production declined significantly.
Mutant strain comparison. Growth, glucose consumption, and alcohol production was quantified in JCL16 (parent), AL1 (first round mutant), and AL2 (second round mutant) during fermentation in shake flasks. Each strain harbored plasmids pIAA11 (alsS–ilvCD) pIAA13 (kivd–ADH2), and pIAA16 (leuA (G462D)–leuBCD)
3MB production and tolerance in AL2
We hypothesized that the cessation of glucose consumption and 3MB production in AL2 could be due to (1) growth/medium conditions or (2) toxicity of the endogenously produced alcohol. The first hypothesis was tested using a richer medium (85 g/L glucose and 10 g/L yeast extract) to increase growth and maintaining the pH at 7 by the addition of 10 M NH4OH as needed. The use of this medium increased growth significantly, as the OD600 was able to reach nearly 10 after just 24 h, compared to the previous high of 4.5. The pH also required constant adjustment and would decrease by ∼1 unit every 12 h. After 36 h, production reached 4.4 g/L with a yield of 0.10 g/g (Fig. 4a). Although this is a significant improvement, just as before, the consumption of glucose and production of 3MB declined shortly after. Next, we began to examine the second hypothesis by testing the sensitivity of AL2 to exogenously added 3MB. Various concentrations of 3MB (between 0 and 5 g/L) were added to cultures of AL2, and the cell viability was measured by counting the number of CFU at 8- and 24-h intervals. A significant growth deficiency was observed at 3MB concentrations as low as 1 g/L (Fig. 5), with severe effects starting at 3 g/L (<17% viability). At 5 g/L, the cell viability was less than 0.1% (Fig. 5). As expected, the toxicity of 3MB is far greater than that of alcohols with a shorter alkyl chain. This finding has previously been demonstrated with 1-butanol and isobutanol in which the tolerance to each of these alcohols is below 16 g/L (Atsumi et al. 2008a; Knoshaug and Zhang 2009), which is significantly lower than that of ethanol (∼50 g/L).
Production of 3MB in AL2. a Growth, glucose consumption, and alcohol production of AL2 (second round mutant) harboring plasmids pIAA11 (alsS–ilvCD), pIAA13 (kivd–ADH2), and pIAA16 (leuA (G462D)–leuBCD) was quantified during fermentation in shake flasks. Filled triangles represent the total 3MB production in AL2 fermentations containing oleyl alcohol for in situ extraction, and open triangles represent the control AL2 fermentations without oleyl alcohol. b Distribution of 3MB and isobutanol in oleyl alcohol fermentations. Filled bars represent the alcohol concentration in the aqueous culture, and open bars represent the alcohol concentration in the oleyl alcohol layer
Two-phase fermentation
These results support the hypothesis that production of 3MB in AL2 is limited by the toxicity of the alcohol as the cell viability at 4.4 g/L is less than 4%. To further examine this hypothesis, we employed a two-phase fermentation system to continuously remove the 3MB from the aqueous cellular environment. It has been shown previously that oleyl alcohol is an effective solvent for the extraction of 1-butanol during fermentation in Clostridium acetobutylicum (Roffler et al. 1987). Therefore, a volume of oleyl alcohol was added to the shake flasks equal to the culture volume at the start of fermentation, creating a two-phase system. Under these conditions, both the cell density and the glucose consumption were significantly higher than the control without oleyl alcohol, and the production of 3MB was able to reach 9.5 g/L after 60 h (Fig. 4a), which is the highest production of any C5 alcohol yet reported in literature. The total yield for 3MB production from glucose was 0.11 g/g. Total alcohol production reached 12.5 g/L, which corresponds to a yield of 0.14 g/g from glucose. Further examination shows that an average of nearly 90% of the total 3MB produced is contained in the oleyl alcohol layer (Fig. 4b). The maximum concentration of 3MB in the aqueous phase reached only 0.8 g/L, at which the cell viability is greater than 70% (Fig. 5). The oleyl alcohol was also able to extract an average of over 81% of the total isobutanol produced, demonstrating that this may be an effective strategy for the extraction of other higher chain alcohols. These results clearly demonstrate that the production of 3MB in AL2 greatly benefits from using a continuous extractive process as the toxicity barrier can be avoided to significantly increase production levels to greater than 5 g/L.
Discussion
A major advantage in the engineering of higher chain alcohol production is the implementation of existing amino acid technologies for process improvement. Of these many technologies, random whole cell mutagenesis and selection with amino acid analogues (Table 1), in particular, has been widely used to develop biocatalysts for the production of many amino acids such as l-threonine, l-isoleucine, l-valine, and l-leucine among many others (Ikeda 2003). Here, we applied this strategy for the development of a strain of E. coli for the production of 3-methyl-1-butanol by selecting for resistance to a l-leucine analogue and screening for increased 3MB production. This strategy is advantageous over rational engineering approaches in that it requires minimal knowledge of the metabolic pathway, and because selective pressure is being used, a large library of mutants can be generated to cover a much more significant portion of sequence space to generate the desired phenotype. Two successive rounds of mutagenesis and selection with AZL resulted in a strain (AL2) able to produce 4.4 g/L of 3MB. This strategy of random mutagenesis and selection can also be used to develop strains for the production of other higher chain alcohols such as isobutanol (l-valine), 1-propanol (l-threonine), 2-methyl-1-butanol (l-isoleucine), and phenylethanol (l-phenylalanine). Furthermore, many of these processes can be challenged with several different analogues in succession, resulting in superior productivity, as oftentimes, many different structural analogues are available for a single amino acid (Table 1). This is also applicable in cases where other amino acids serve as precursors to the final amino acid of interest. For example, 2-methyl-1-butanol is synthesized from an l-isoleucine precursor which itself is synthesized from l-threonine. Therefore, a strain can be developed using structural analogues for both l-threonine (α-amino-β-hydroxyvaleric acid) and l-isoleucine (d,l-4-thiaisoleucine; Table 1).
Analysis of the host tolerance to 3MB revealed that the productivity in this strain may be limited by the host’s sensitivity to the product. The viability of AL2 at 4.4 g/L of 3MB was found to be less than 4%, suggesting that the toxicity of the product is limiting its own production and growth. A two-phase fermentation with oleyl alcohol allowed for a prolonged production phase in which 3MB was able to reach a total of 9.5 g/L after 60 h with a yield of 0.11 g/g, with nearly 90% of the 3MB contained within the oleyl alcohol. The yield of this experiment is 33% of the theoretical maximum (0.33 g/g), which is calculated from a stoichiometric balance from glucose to 3MB under the assumption that excess reducing power in the form of NAD(P)H can be recycled to NAD(P)+ by O2. Total alcohol production reached 12.5 g/L. The success of this approach increases the potential of producing 3MB as a biofuel from E. coli as titers can potentially reach in excess of 30 g/L before the aqueous concentration of 3MB again becomes troublesome.
This work also demonstrates the utility and potential in using E. coli as a host for biofuel production. The accumulated wealth of knowledge and developed genetic tools, as well as previous successes in pathway engineering and metabolite production (Chao et al. 1993; Farmer and Liao 2001; Martin et al. 2003) as well as amino acid production (Ikeda 2003), make E. coli an attractive host for higher chain alcohol production via the 2-keto acid pathways. As production titers continue to increase, existing tools and technology for product removal (Roffler et al. 1987) and increasing host solvent tolerance (Alper et al. 2006; Atsumi et al. 2008a; Borden and Papoutsakis 2007; Yomano et al. 1998), along with the elucidation of solvent response pathways (Brynildsen and Liao 2009; Tomas et al. 2004), can be applied to maintain the development of these processes.
References
Alper H, Moxley J, Nevoigt E, Fink GR, Stephanopoulos G (2006) Engineering yeast transcription machinery for improved ethanol tolerance and production. Science 314:1565–1568
Atsumi S, Liao JC (2008) Metabolic engineering for advanced biofuels production from Escherichia coli. Curr Opin Biotechnol 19:414–419
Atsumi S, Hanai T, Liao JC (2008a) Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature 451:86–89
Atsumi S, Cann AF, Connor MR, Shen CR, Smith KM, Brynildsen MP, Chou KJ, Hanai T, Liao JC (2008b) Metabolic engineering of Escherichia coli for 1-butanol production. Metab Eng 10:305–311
Borden JR, Papoutsakis ET (2007) Dynamics of genomic-library enrichment and identification of solvent tolerance genes for Clostridium acetobutylicum. Appl Environ Microbiol 73:3061–3068
Brynildsen MP, Liao JC (2009) An integrated network approach identifies the isobutanol response network of Escherichia coli. Mol Syst Biol 5:277
Cann AF, Liao JC (2008) Production of 2-methyl-1-butanol in engineered Escherichia coli. Appl Microbiol Biotechnol 81:89–98
Chao YP, Patnaik R, Roof WD, Young RF, Liao JC (1993) Control of gluconeogenic growth by pps and pck in Escherichia coli. J Bacteriol 175:6939–6944
Connor MR, Liao JC (2008) Engineering of an Escherichia coli strain for the production of 3-methyl-1-butanol. Appl Environ Microbiol 74:5769–5775
Connor MR, Liao JC (2009) Microbial production of advanced transportation fuels in non-natural hosts. Curr Opin Biotechnol 20:307–315
Dinneen B (2008) Changing the climate: Ethanol Industry Outlook 2008. In: RFA outlook. Renewable Fuels Association. http://www.ethanolrfa.org/objects/pdf/outlook/RFA_Outlook_2008.pdf. Accessed October 6, 2009
Farmer WR, Liao JC (2001) Precursor balancing for metabolic engineering of lycopene production in Escherichia coli. Biotechnol Prog 17:57–61
Gusyatiner MM, Lunts MG, Kozlov YI, Ivanovskaya LV, Voroshilova EB (2002) DNA coding for mutant isopropylmalate synthase l-leucine-producing microorganism and method for producing l-leucine. United States Patent 6,403,342
Ikeda M (2003) Amino acid production processes. In: Scheper T (ed) Advances in biochemical engineering/technology. Springer, Berlin, pp 1–35
Kim Y, Ingram LO, Shanmugam KT (2007) Construction of an Escherichia coli K-12 mutant for homoethanologenic fermentation of glucose or xylose without foreign genes. Appl Environ Microbiol 73:1766–1771
Knoshaug EP, Zhang M (2009) Butanol tolerance in a selection of microorganisms. Appl Biochem Biotechnol 153:13–20
Lee SY, Park JH, Jang SH, Nielsen LK, Kim J, Jung KS (2008) Fermentative butanol production by Clostridia. Biotechnol Bioeng 101:209–228
Lee JY, Jang YS, Lee J, Papoutsakis ET, Lee SY (2009) Metabolic engineering of Clostridium acetobutylicum M5 for highly selective butanol production. Biotechnol J 4:1432–1440
Martin VJ, Pitera DJ, Withers ST, Newman JD, Keasling JD (2003) Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nat Biotechnol 21:796–802
Miller JH (1972) Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor
Roffler SR, Blanch HW, Wilke CR (1987) In-situ recovery of butanol during fermentation. Bioprocess Biosyst Eng 2:1–12
Seiferlein KE (2009) Annual energy review 2008. In: Annual energy review. Energy Information Administration. http://www.eia.doe.gov/emeu/aer/contents.html. Accessed October 6, 2009
Shen CR, Liao JC (2008) Metabolic engineering of Escherichia coli for 1-butanol and 1-propanol production via the keto-acid pathways. Metab Eng 10:312–320
Stephanopoulos G (2007) Challenges in engineering microbes for biofuels production. Science 315:801–804
Tomas CA, Beamish J, Papoutsakis ET (2004) Transcriptional analysis of butanol stress and tolerance in Clostridium acetobutylicum. J Bacteriol 186:2006–2018
Withers ST, Gottlieb SS, Lieu B, Newman JD, Keasling JD (2007) Identification of isopentenol biosynthetic genes from Bacillus subtilis by a screening method based on isoprenoid precursor toxicity. Appl Environ Microbiol 73:6277–6283
Yan Y, Liao JC (2009) Engineering metabolic systems for production of advanced fuels. J Ind Microbiol Biotechnol 36:471–479
Yomano LP, York SW, Ingram LO (1998) Isolation and characterization of ethanol-tolerant mutants of Escherichia coli KO11 for fuel ethanol production. J Ind Microbiol Biotechnol 20:132–138
Zhang K, Sawaya MR, Eisenberg DS, Liao JC (2008) Expanding metabolism for biosynthesis of nonnatural alcohols. Proc Natl Acad Sci U S A 105:20653–20658
Acknowledgments
We would also like to thank the UCLA-DOE Joint Genome Institute and Gevo, Inc. for providing funding for this work.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Connor, M.R., Cann, A.F. & Liao, J.C. 3-Methyl-1-butanol production in Escherichia coli: random mutagenesis and two-phase fermentation. Appl Microbiol Biotechnol 86, 1155–1164 (2010). https://doi.org/10.1007/s00253-009-2401-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-009-2401-1