Abstract
Novel dissimilatory perchlorate-reducing bacteria (DPRB) were isolated from enrichments conducted under conditions different from those of all previously described DPRB. Strain LT-1T was enriched using medium buffered at pH 6.6 with 2-(N-morpholino)ethanesulfonic acid (MES) and had only 95% 16S rRNA gene identity with its closest relative, Azonexus caeni. Strain MPT was enriched in the cathodic chamber of a perchlorate-reducing bioelectrical reactor (BER) and together with an additional strain, CR (99% 16S rRNA gene identity), had 97% 16S rRNA gene identity with Propionivibrio limicola. The use of perchlorate and other electron acceptors distinguished strains MPT and CR from P. limicola physiologically. Strain LT-1T had differences in electron donor utilization and optimum growth temperatures from A. caeni. Strains LT-1T and MPT are the first DPRB to be described in the Betaproteobacteria outside of the Dechloromonas and Azospira genera. On the basis of phylogenetic and physiological features, strain LT-1T represents a novel genus in the Rhodocyclaceae; strain MPT represents a novel species within the genus Propionivibrio. The names Dechlorobacter hydrogenophilus gen. nov., sp. nov and Propionivibrio militaris sp. nov. are proposed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Perchlorate has been widely used as an oxidant in munitions propellant as well as other applications and, due to its competitive inhibition of iodine uptake in the thyroid gland, can cause decreased levels of thyroid hormone production (Wolff 1998). As a result of unregulated disposal of the compound prior to 1997, perchlorate contamination is now widespread (Coates and Achenbach 2004). DPRB are capable of perchlorate reduction completely to harmless chloride and therefore represent an ideal means for remediating this compound (Coates and Achenbach 2004). DPRB have been isolated from a variety of locales, and perchlorate reduction genes have been found in areas as remote as Antarctica (Bender et al. 2004; Coates et al. 1999). Their existence in such varied, and sometimes pristine, habitats has been puzzling in the context of DPRB research primarily focused on bioremediation of a contaminant perceived as anthropogenic. Recently, however, several reports have discovered natural perchlorate deposition in a variety of locales, indicating a potential natural source, which, if confirmed, would help explain the ubiquity of these organisms (Rajagopalan et al. 2006, 2009; Rao et al. 2007; Scanlon et al. 2008).
In spite of their broad geographic distribution, DPRB have been phylogenetically limited to the Proteobacteria (Coates and Achenbach 2004). Thus far, organisms in pure culture occur in only five genera in the Alpha-, Beta-, and Epsilonproteobacteria classes. It is possible that perchlorate reduction may have only evolved in a common ancestor within the Proteobacteria and was subsequently lost in multiple genera, but a more parsimonious theory is that perchlorate reduction evolved recently and has been transferred horizontally. This theory is supported by a lack of phylogenetic synteny between the 16S rRNA genes and chlorite dismutase (a required perchlorate-reduction pathway gene) genes from the same organisms (Bender et al. 2004). The limited phylogenetic representation of DPRB given their ubiquity and the probability of horizontal gene transfer makes little sense without considering culture bias. Although a few of these organisms have been isolated with hydrogen as an electron donor (Miller and Logan 2000; Nerenberg et al. 2002, 2006; Shrout et al. 2005; Zhang et al. 2002), most have been isolated under mesophilic, chemoorganoheterotrophic conditions (Bruce et al. 1999; Coates et al. 1999, 2001; Horn et al. 2005; Michaelidou et al. 2000; Rikken et al. 1996; Wallace et al. 1996; Wolterink et al. 2005).
A variety of studies have shown that even with the same starting inoculum, alteration of enrichment conditions can lead to isolation of novel organisms, both physiologically and phylogenetically (Auman et al. 2000; Bussmann et al. 2004; Connon and Giovannoni 2002; Grosser et al. 2000; Orphan et al. 2000). The isolation of two DPRB from the cathodic chamber of a bioelectrical reactor (BER) confirmed this, since no perchlorate-reducing organisms had been previously enriched under such circumstances (Thrash et al. 2007). Dechlorospirillum sp. strain VDY had novel physiological characteristics compared to other tested DPRB (Thrash et al. 2007). Furthermore, enrichments carried out at 55°C lead to the isolation of the first thermophilic microorganism capable of perchlorate reduction (Balk et al. 2008), although the presence of known perchlorate-reduction pathway genes was not confirmed and clear evidence that growth was dependent on perchlorate was not provided, so it is unknown whether or not this organism was a dissimilatory or co-incidental perchlorate reducer. Regardless, Moorella perchloratireducens was also the first bacterium isolated outside of the Proteobacteria capable of perchlorate reduction, providing additional evidence that alteration of enrichment conditions leads to novel physiologies and phylogenetic affiliations and sometimes both in the same organism.
In the course of our continuing studies on the diversity of organisms capable of dissimilatory perchlorate reduction, three new isolates have been characterized and are reported here, two of which were enriched using conditions that were unique compared to that of previously described DPRB. Strain MPT, the other organism isolated from the cathodic chamber of the same BER enrichment (Thrash et al. 2007), its closest relative, strain CR, and strain LT-1T were mesophilic, strictly respiratory, facultative anaerobes that were phylogenetically distinct from all other previously described DPRB. Strains MPT and CR are the first DPRB to be identified in the genus Propionivibrio, and strain LT-1T is the first from the new genus, proposed here as Dechlorobacter gen. nov. The unique phylogenetic affiliation of these strains supports previous observations that alteration of enrichment and isolation conditions can select for unique organisms capable of perchlorate reduction. The environmental role of these new species in the natural redox cycle of chlorine is still unknown. However, the results of these studies do indicate that the true phylogenetic diversity of organisms capable of perchlorate reduction still remains to be discovered.
Materials and methods
Culture conditions, enrichments, and isolation
Unless otherwise mentioned, the basal medium used for cultures was the same as previously described (Bruce et al. 1999; Thrash et al. 2007). Strain MPT was enriched as previously described (Thrash et al. 2007). One-milliliter samples from the cathodic chamber were used to inoculate basal bicarbonate medium containing 5 mM anthrahydroquinone-2,6-disulfonate (AHDS) and 10 mM perchlorate amended with 0.1 mM acetate as a suitable carbon source. Medium showing growth was then streaked onto R2A (Difco, BD) plates. A single white colony was picked and transferred to liquid medium containing 10 mM acetate and perchlorate. Strains CR and strain LT-1T were isolated using the standard shake tube method previously described (Bruce et al. 1999). For determining pH-dependent growth characteristics, anoxic phosphate-buffered medium was used. NaH2PO4 and Na2HPO4 were added in appropriate concentrations to establish stable pH at 6.0, 6.5, 6.8, 7.0, 7.2, and 7.5. These were boiled, cooled, and dispensed under a 100% N2 headspace. Omitting sodium bicarbonate, all other medium components were identical with bicarbonate-buffered medium. Cultures were grown in triplicate at each different pH and monitored using optical density at 600 nm. Electron donors and acceptors were added from sterile anoxic stocks. Alternative electron acceptors were tested in media amended with 10 mM acetate and alternative electron donors with 10 mM perchlorate. For the determination of alternative electron acceptors and donors, substrate utilization was judged positive only after a minimum of three consecutive transfers under growth conditions.
Analytical methods
All experimental analyses were performed in triplicate to ensure reproducibility and the results are expressed as the mean of these determinations. The concentration of perchlorate and other anions in cultures was determined using ion chromatography as previously described (Chaudhuri et al. 2002). Fe(II) concentration was determined using the Ferrozine assay (Lovley and Phillips 1987). Culture purity was verified by microscopic inspection and 16S rRNA gene sequence analysis (below). Cell density increase was monitored by optical density changes at 600 nm (OD600).
Microscopy
Cells were observed using phase contrast microscopy on a Zeiss Axioskop 2 plus. Scanning electron microscopy was carried out as described (Weber et al. 2009) by harvesting cells, washing twice with 0.1 M sodium cacodylate buffer pH 7.2, and fixing with glutaraldehyde. Cells were then resuspended in 1% osmium tetroxide in sodium cacodylate buffer for 2 h and rinsed in sodium cacodylate buffer. Cells were then dehydrated for 10 min in 35%, 50%, 70%, 80%, 95%, and 100% ethanol, followed by critical point drying, mounting onto stubs and sputter coated with palladium/gold, and viewing with a Hitachi S5000 scanning electron microscope at 20 kV.
Whole-cell fatty acid analysis
Whole-cell fatty acid content was determined by growing cells in basal medium as described above, harvesting by centrifugation, and re-suspending in 1 mL buffer. Fatty acids were identified using the Sherlock Microbial Identification System (v. 4.5, MIDI, Newark, DE) according to the manufacturer’s protocol.
Molecular/phylogenetic analysis
Genomic DNA of strain MPT was isolated from pure cultures using the Power Soil Kit (MoBio, Carlsbad, CA) according to the manufacturer’s protocol and from strains CR and LT-1T as previously described (Thrash et al. 2007). 16S rRNA gene PCR on genomic DNA from strain MPT was carried out as described previously (Wrighton et al. 2008) and from strains CR and LT-1T as described previously elsewhere (Thrash et al. 2007). PCR of the cld gene sequences was carried out as described by Bender et al. (Bender et al. 2004). 16S rRNA gene sequences were aligned with MUSCLE 3.6 (Edgar 2004) and analyzed with MrBayes 3.2 (Huelsenbeck and Ronquist 2001; Ronquist and Huelsenbeck 2003) using four chains until the standard deviation of the split frequencies was stabilized below 0.01, in this case for 1,158,000 generations, with a sample frequency of 1,000. The first 25% of the samples were discarded for accurate estimation of the posterior probability distribution of the summary tree.
Results
Both strains MPT and LT-1T were enriched under unusual conditions compared with previously described DPRB. Strain MPT was enriched in the cathodic chamber of a BER in the presence of the electron shuttle anthroquinone-2,6-disulfonate (Thrash et al. 2007) and isolated by inoculating medium containing 10 mM acetate and perchlorate, prior to streaking onto aerobic R2A plates after growth was observed. A single colony was picked in an anaerobic chamber and transferred to anoxic medium containing 10 mM acetate and perchlorate. Strain LT-1T was enriched using slightly acidic (pH 6.6) 2-(N-morpholino)ethanesulfonic acid (MES)-buffered medium inoculated with soil from the Longhorn Army Ammunition Plant, TX. Strain CR was enriched as previously described (Coates et al. 1999) with perchlorate-contaminated soil from Los Alamos National Laboratory as the inoculum.
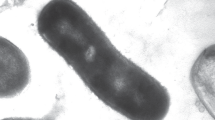
Strains MPT and CR grew singly as curved rods, approximately 2 µm long × 0.3 µm in diameter (Fig. 1). Strain LT-1T grew as a slightly curved (Fig. 2a) or short, straight rod (Fig. 2b), 0.8–1.6 × 0.3 µm, either singly or in chains. Cells from all three strains were observed by phase-contrast microscopy to be motile when grown on acetate and perchlorate (10 mM). MPT, CR, and LT-1T were strictly respiratory, as judged by their inability to grow in the absence of an electron acceptor either with 10 mM glucose, or in rich media (10 mM glucose, 1 g/L casamino acids, and 1 g/L yeast extract). None of the strains were capable of growth after pasteurization at 80°C and were therefore considered incapable of forming spores. All three strains grew by complete reduction of perchlorate to chloride, and all three strains contained copies of the chlorite dismutase gene (cld) (Bender et al. 2004), previously reported for strains LT-1T and CR and confirmed in this study for strain MPT (data not shown). The cld sequence for strain MPT was most closely related to that of CR. Figure 3 shows typical growth curves for strain MPT (Fig. 3a) and strain LT-1T (Fig. 3b) on 10 mM acetate and perchlorate. Cell density increase was monitored by optical density changes at 600 nm. Strains MPT and CR had maximum growth rates at 30°C of 0.21 and 0.22 h−1, respectively. In addition, for both strains, chlorate accumulated to ∼2 mM (2.1 mM for strain MPT—Fig. 2) when grown on 10 mM perchlorate. In contrast, strain LT-1T showed much less (maximum concentration ∼0.1 mM) and more transient chlorate accumulation (Fig. 3b). Strain LT-1T had a maximum growth rate of 0.28 h−1 at 37°C.
Growth curves of strain MPT (a) and strain LT-1T (b). Cell number increase was monitored with optical density (OD) at 600 nm. Closed circles OD, open squares [ClO −4 ], open triangles [Cl−], open diamonds, [ClO −3 ]; all concentrations in millimolar. Error bars represent the standard deviation of triplicate experiments
Table 1 describes the comparable substrate utilization range for the three strains. In general, they reduced a similar suite of electron acceptors and utilized similar electron donors. Strain CR was not capable of utilizing Fe(II) (as FeCl2) or H2, however, and exhibited a lower NaCl tolerance. It was capable of growing at 40°C, whereas strain MPT could not, and strain CR grew optimally at pH 7 as compared to 6.8 for strain MPT. LT-1T grew optimally at pH 6.5, which is unusual for DPRB, but is not unexpected since it was enriched in MES-buffered medium at pH 6.6. However, maximum growth rates in MES-buffered medium were considerably lower than those on bicarbonate-buffered medium (0.03 vs. 0.16 h−1 at 37°C, respectively).
Fatty acid composition for strains MPT, CR, and LT-1T are shown in Table 2. The primary dominant fatty acid for strain MPT was the summed feature 16:1ω7c/15 iso 2OH; for CR and LT-1T, it was 16:1c9. 16:0 was the second most abundant fatty acid in all three strains.
Analysis of the 16S rRNA gene sequences indicated that all three strains were members of the Rhodocyclaceae family in the Betaproteobacteria (Fig. 4). Sequences for strains MPT and CR were 99% identical, and both strains were most closely related to Propionivibrio limicola GolChi1T (97% identity), making them probable members of the Propionivibrio genus. Strain LT-1T was most closely related to the Azonexus genus, having 95% identity with Azonexus caeni.
Discussion
These studies resulted in the isolation and characterization of three novel organisms that lie outside of the phylogenetic groupings of known DPRB. In spite of the close identity to P. limicola, both strains MPT and CR had only 95% identity to Propionivibrio decarboxylicus and Propionivibrio pelophilus, the other described species in the genus. Furthermore, while all members of the Propionivibrio were fermentative and formed acetate and propionate as end products (Brune et al. 2002; Hippe et al. 1999; Meijer et al. 1999; Tanaka et al. 1990), strains MPT and CR were strictly respiratory and completely oxidized acetate to CO2. P. limicola was incapable of utilizing external electron acceptors (oxygen, nitrate, thiosulfate, sulfur, and amorphous ferric iron), whereas strains MPT and CR utilized oxygen, nitrate, chlorate, and perchlorate. Furthermore, P. limicola also was unable to make use of carboxylic acids, whereas MPT and CR could use many (Table 1). It is unknown whether or not P. limicola can reduce perchlorate, but since the perchlorate reduction pathway involves production of oxygen and subsequent reduction of it by cytochrome oxidase (Achenbach et al. 2006; Rikken et al. 1996) and the pcrC gene encodes for a c-type cytochrome (Bender et al. 2005), it is unlikely that this organism could make use of perchlorate since it cannot reduce oxygen and contains no cytochromes (Brune et al. 2002). Strains MPT and CR were also distinct from P. limicola morphologically and were curved rods, whereas P. limicola is a straight rod (Brune et al. 2002). Given the combination of physiological and morphological distinction with the phylogenetic distance from P. limicola, strain MPT should be considered a new species, with the proposed name Propionivibrio militaris sp. nov. Given the significant similarity to strain MPT, CR is considered a strain of P. militaris.
Strain LT-1T had only 95% 16S rRNA gene identity to its nearest relative, A. caeni, which distinguished it as a novel genus. In addition, strain LT-1T was morphologically distinct from both A. caeni and A. fungiphilus, the two described Azonexus species. LT-1T was a short, straight rod, whereas A. caeni was more rounded and asymmetrical (Quan et al. 2006), and A. fungiphilus was a long, curved rod, which sometimes was capable of growing filamentously (Reinhold-Hurek and Hurek 2000). Strain LT-1T was also smaller than either of the Azonexus species. Physiologically, strain LT-1T was distinct from A. caeni in that it could use perchlorate, chlorate, and Mn(IV) as terminal electron acceptors and butyrate, iso-valerate, fumarate, hexanoic acids, ethanol, H2, and AHDS as electron donors; A. caeni could not use any of these organic electron donors (Quan et al. 2006). A. fungiphilus was an obligate aerobe (Reinhold-Hurek and Hurek 2000), and A. caeni was a facultative anaerobe that was capable of denitrification (Quan et al. 2006). It is unknown if these organisms could use perchlorate. A. caeni had a growth optimum at between 25°C and 30°C, whereas strain LT-1T had a growth optimum at 37°C. Taken together, the phylogenetic, morphological, and physiological differences between strain LT-1T and A. caeni indicate that strain LT-1T represents a novel species of a new genus, and the name Dechlorobacter hydrogenophilus is proposed.
Chlorate accumulation in DPRB is not well understood. Strains MPT and LT-1T accumulated chlorate in much different amounts (Fig. 3). However another DPRB, Azospira sp. strain PCC, had comparable chlorate accumulation to that of strains MPT and CR as a result of higher relative affinity for perchlorate versus chlorate, making perchlorate a competitive inhibitor of chlorate reduction (Dudley et al. 2008). A reasonable hypothesis is that the perchlorate reduction genes in MPT and CR are more similar to those of PCC than to those of strain LT-1T and other DPRB that do not exhibit this type of intermediate accumulation, but this needs to be tested.
The new appreciation for naturally occurring perchlorate brought about by the work of Rajagopalan et al. (2006, 2009), Rao et al. (2007), and Scanlon et al. (2008) makes the search for organisms capable of using this compound for respiration much more than a means to understand and increase the operational parameters of perchlorate bioremediation schemes. Although DPRB have been isolated from a variety of locales, the natural abundance, diversity, and ecology of DPRB has yet to be explored. The new evidence of perchlorate as a naturally occurring compound makes the study of this metabolism important in understanding the biological processes involved in natural cycling of chlorine. Current isolates will be indispensable for understanding the physiology behind future culture-independent studies. Recent evidence for perchlorate on Mars (Hecht et al. 2009a, b; Mumma et al. 2009) even presents the possibility of studying chemolithoautotrophic DPRB as model organisms for Martian astrobiology, and comparative cell biology will only be available with multiple organisms capable of these metabolisms.
However, in spite of the new horizons for examining DPRB in the environment, studies continue to demonstrate the prevalence of perchlorate as a contaminant and one that has made its way to the top of the food chain, having been found in dairy and breast milk (Dasgupta et al. 2008; Dyke et al. 2007; Kirk et al. 2005). As a result, the need for effective remediation technology for perchlorate continues to grow. With bioremediation as the most effective means to completely eliminate this compound, the development and testing of new strategies will most likely lead to the discovery of novel DPRB, since new treatment processes can lead to new enrichment conditions. BER technology (Thrash et al. 2007) and hydrogen-based reactors (Miller and Logan 2000; Nerenberg et al. 2002) are examples of treatment alternatives that have already enriched for novel organisms. A remarkable number of BER options exist (Thrash and Coates 2008), and new reactors using these as well as the application of microbial fuel cell technology (already shown capable of stimulating nitrate-reducing microorganisms (Clauwaert et al. 2007)) for perchlorate bioremediation (Shea et al. 2008) should lead to even more disparate isolates.
Description of Dechlorobacter gen. nov.
Dechlorobacter [de.chlor.o.bac’ter L. pref. de from; Gr. adj. chloros green (chlorine); N.L. masc. n. bacter a short rod; N.L. masc. n. Dechlorobacter dechlorinating rod].
Rod-shaped cells, 0.8–1.6 × 0.3 µm, non-spore-forming, non-fermentative, facultative anaerobe. Cells are motile and occur singly or in chains. Gram negative. Strictly respiring, complete oxidizer that oxidizes acetate with O2, ClO −4 , ClO −3 , Mn(IV), or NO −3 as electron acceptors. Perchlorate and chlorate are completely reduced to Cl−. Type species is D. hydrogenophilus.
Description of Dechlorobacter hydrogenophilus sp. nov.
D. hydrogenophilus [hy.dro.gen’o.phi’lus L. n. hydrogen the dihydrogen molecule H2; phi’lus L. v. loving hydrogenophilus loving hydrogen, on which it can grow].
Optimum growth occurs at 37°C, pH 6.5, in freshwater basal medium with 0% NaCl. Cells can grow over the temperature range 4–37°C but not at 42°C and between pH 6.0–7.2. Cells can use organic substrates such as acetate and malate and can grow mixotrophically with hydrogen as the electron donor. Cells could not utilize formate, lactate, citrate, heptanoic acid, Fe(II), thiosulfate, fumarate, ethanol, methanol, glycerol, glucose, lactose, sucrose, fructose, maltose, ribose, benzene, benzoate, toluene, p-Cresol, phenol, phenanthrene, or octane as electron donors and could not utilize sulfate, sulfur, ferric citrate, ferric nitrilotriacetate (NTA), ferric pyrophosphate, fumarate, or selenate as electron acceptors. Cells contain the fatty acids 10:0 3OH, 12:0, 12:0 3OH, 14:0, 16:1c9, 16:0, and 18:1c11/t9/t6. Type species is D. hydrogenophilus. The type strain is LT-1T (ATCC BAA-1869) and was isolated from soil collected at the Longhorn Army Ammunition Plant, Longhorn, TX.
Description of Propionivibrio militaris sp. nov.
P. militaris [mil’i.tar.is L. adj. militaris martial, war-like, referring to the ability of the organism to degrade perchlorate, frequently used in rocket propellants].
Curved rod-shaped cells, ∼2 × 0.3 µm, non-spore-forming, non-fermentative, facultative anaerobe. Gram negative. Cells are motile and occur singly. Strictly respiring, complete oxidizer that oxidizes acetate with O2, ClO −4 , ClO −3 , NO −3 or NO −2 as electron acceptors. Perchlorate and chlorate are completely reduced to Cl− with chlorate accumulation during growth on perchlorate. Cells can use chemoorganotrophic substrates such as acetate and propionate as electron donors for growth as well as AHDS. Cells could not use formate, methanol, ethanol, catechol, glycerol, benzoate, citrate, glucose, sucrose, fructose, maltose, or urea as electron donors and could not utilize sulfate, thiosulfate, fumarate, malate, ferric NTA, or AQDS as electron acceptors. Optimum growth occurs at 30°C in freshwater basal medium with 0% NaCl. Cells contain the fatty acids 10:0 3OH, 12:0, and 16:0. Type species is P. militaris. The type strain is MPT (DSM 21683, ATCC BAA-1728) and was isolated from the cathodic chamber of an active perchlorate-reducing BER enrichment inoculated with water from Strawberry Creek at UC Berkeley, Berkeley, CA.
The type strain, MPT, has the following additional characteristics: Cells can utilize the inorganic electron donors hydrogen and Fe(II). Cells grow between 10°C and 37°C, but not at 40°C. Growth occurs at pH 6.0–7.5, with 6.8 optimum. Cells also contain the fatty acids 16:1ω7c/15 iso 2OH, 17:0 cyclo, and 18:1ω7c.
References
Achenbach LA, Bender KS, Sun Y, Coates JD (2006) The biochemistry and genetics of microbial perchlorate reduction. In: Gu B, Coates JD (eds) Perchlorate: environmental occurrence, interactions and treatment. Springer, New York, pp 297–310
Auman AJ, Stolyar S, Costello AM, Lidstrom ME (2000) Molecular characterization of methanotrophic isolates from freshwater lake sediment. Appl Environ Microbiol 66:5259–5266
Balk M, van Gelder T, Weelink SA, Stams AJM (2008) (Per)chlorate reduction by the thermophilic bacterium Moorella perchloratireducens sp. nov., isolated from underground gas storage. Appl Environ Microbiol 74:403–409
Bender KS, Rice MR, Fugate WH, Coates JD, Achenbach LA (2004) Metabolic primers for detection of (Per)chlorate-reducing bacteria in the environment and phylogenetic analysis of cld gene sequences. Appl Environ Microbiol 70:5651–5658
Bender KS, Shang C, Chakraborty R, Belchik SM, Coates JD, Achenbach LA (2005) Identification, characterization, and classification of genes encoding perchlorate reductase. J Bacteriol 187:5090–5096
Bruce RA, Achenbach LA, Coates JD (1999) Reduction of (per)chlorate by a novel organism isolated from a paper mill waste. Environ Microbiol 1:319–331
Brune A, Ludwig W, Schink B (2002) Propionivibrio limicola sp. nov., a fermentative bacterium specialized in the degradation of hydroaromatic compounds, reclassification of Propionibacter pelophilus as Propionivibrio pelophilus comb. nov. and amended description of the genus Propionivibrio. Int J Syst Bacteriol 52:441–444
Bussmann I, Pester M, Brune A, Schink B (2004) Preferential cultivation of type II methanotrophic bacteria from littoral sediments (Lake Constance). FEMS Microbiol Ecol 47:179–189
Chaudhuri SK, O’Connor SM, Gustavson RL, Achenbach LA, Coates JD (2002) Environmental factors that control microbial perchlorate reduction. Appl Environ Microbiol 68:4425–4430
Clauwaert P, Rabaey K, Aelterman P, DeSchamphelaire L, Pham TH, Boeckx P, Boon N, Verstraete W (2007) Biological denitrification in microbial fuel cells. Environ Sci Technol 41:3354–3360
Coates JD, Achenbach LA (2004) Microbial perchlorate reduction: rocket-fuelled metabolism. Nat Rev Microbiol 2:569–580
Coates JD, Michaelidou U, Bruce RA, O’Connor SM, Crespi JN, Achenbach LA (1999) Ubiquity and diversity of dissimilatory (Per)chlorate-reducing bacteria. Appl Environ Microbiol 65:5234–5241
Coates JD, Chakraborty R, Lack JG, O’Connor SM, Cole KA, Bender KS, Achenbach LA (2001) Anaerobic benzene oxidation coupled to nitrate reduction in pure culture by two strains of Dechloromonas. Nature 411:1039–1043
Connon S, Giovannoni SJ (2002) High-throughput methods for culturing microorganisms in very-low-nutrient media yield diverse new marine isolates. Appl Environ Microbiol 68:3878–3885
Dasgupta PK, Kirk AB, Dyke JV, Ohira S-I (2008) Intake of iodine and perchlorate and excretion in human milk. Environ Sci Technol 42:8115–8121
Dudley M, Salmone A, Nerenberg R (2008) Kinetics of a chlorate-accumulating, perchlorate-reducing bacterium. Water Res 42:2403–2410
Dyke JV, Ito K, Obitsu T, Hisamatsu Y, Dasgupta PK, Blount BC (2007) Perchlorate in dairy milk. Comparison of Japan versus the United States. Environ Sci Technol 41:88–92
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797
Grosser RJ, Friedrich M, Ward DM, Inskeep WP (2000) Effect of model sorptive phases on phenanthrene biodegradation: different enrichment conditions Influence bioavailability and selection of phenanthrene-degrading isolates. Appl Environ Microbiol 66:2695–2702
Hecht MH, Catling DC, Clark BC, DeFlores L, Gospodinova K, Kapit J, Kounaves SP, Ming DW, Quinn RC, West SJ, Young SMM (2009a) Perchlorate in martian soil: evidence and implications, 40th lunar and planetary science conference. Lunar and Planetary Institute, The Woodlands
Hecht MH, Kounaves SP, Quinn RC, West SJ, Young SMM, Ming DW, Catling DC, Clark BC, Boynton WV, Hoffman J, DeFlores LP, Gospodinova K, Kapit J, Smith PH (2009b) Detection of perchlorate and the soluble chemistry of martian soil at the phoenix lander site. Science 325:64–67
Hippe H, Hagelstein A, Kramer I, Swiderski J, Stackebrandt E (1999) Phylogenetic analysis of Formivibrio citricus, Propionivibrio dicarboxylicus, Anaerobiospirillum thomasii, Succinimonas amylolytica and Succinivibrio dextrinosolvens and proposal of Succinivibrionaceae fam. nov. Int J Syst Evol Microbiol 49:779–782
Horn MA, Ihssen J, Matthies C, Schramm A, Acker G, Drake HL (2005) Dechloromonas denitrificans sp. nov., Flavobacterium denitrificans sp. nov., Paenibacillus anaericanus sp. nov., and Paenibacillus terrae strain MH72, N2O-producing bacteria isolated from the gut of the earthworm Aporrectodea caliginosa. Int J Syst Evol Microbiol 55:1255–1265
Huelsenbeck JP, Ronquist F (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755
Kirk AB, Martinelango PK, Tian K, Dutta A, Smith EE, Dasgupta PK (2005) Perchlorate and iodide in dairy and breast milk. Environ Sci Technol 39:2011–2017
Lovley DR, Phillips EJ (1987) Rapid assay for microbially reducible ferric iron in aquatic sediments. Appl Environ Microbiol 53:1536–1540
Meijer WG, Nienhuis-Kuiper ME, Hansen TA (1999) Fermentative bacteria from estuarine mud: phyIogenetic position of Acidaminobacter hydrogenoformans and description of a new type of Gram-negative, propionigenic bacterium as Propionibacter pelophilus gen. nov., sp. nov. Int J Syst Evol Microbiol 49:1039–1044
Michaelidou U, Achenbach LA, Coates JD (2000) Isolation and characterization of two novel (Per)chlorate-reducing bacteria from swine waste lagoons, vol 57. Kluwer, New York
Miller JP, Logan BE (2000) Sustained perchlorate degradation in an autotrophic, gas-phase, packed-bed bioreactor. Environ Sci Technol 34:3018–3022
Mumma MJ, Villanueva GL, Novak RE, Hewagama T, Bonev BP, DiSanti MA, Mandell AM, Smith MD (2009) Strong release of methane on mars in northern summer 2003. Science 332:1041–1045
Nerenberg R, Rittmann BE, Najm I (2002) Perchlorate reduction in a hydrogen-based membrane-biofilm reactor. J Am Water Works Assoc 94:103–114
Nerenberg R, Kawagoshi Y, Rittmann BE (2006) Kinetics of a hydrogen-oxidizing, perchlorate-reducing bacterium. Water Res 40:3290–3296
Orphan VJ, Taylor LT, Hafenbradl D, DeLong EF (2000) Culture-dependent and culture-independent characterization of microbial assemblages associated with high-temperature petroleum reservoirs. Appl Environ Microbiol 66:700–711
Quan Z-X, Im W-T, Lee S-T (2006) Azonexus caeni sp. nov., a denitrifying bacterium isolated from sludge of a wastewater treatment plant. Int J Syst Evol Microbiol 56:1043–1046
Rajagopalan S, Anderson TA, Fahlquist L, Rainwater KA, Ridley M, Jackson WA (2006) Widespread presence of naturally occurring perchlorate in high plains of Texas and New Mexico. Environ Sci Technol 40:3156–3162
Rajagopalan S, Anderson T, Cox S, Harvey G, Cheng Q, Jackson WA (2009) Perchlorate in wet deposition across North America. Environ Sci Technol 43:616–622
Rao B, Anderson TA, Orris GJ, Rainwater KA, Rajagopalan S, Sandvig RM, Scanlon BR, Stonestrom DA, Walvoord MA, Jackson WA (2007) Widespread natural perchlorate in unsaturated zones of the Southwest United States. Environ Sci Technol 41:4522–4528
Reinhold-Hurek B, Hurek T (2000) Reassessment of the taxonomic structure of the diazotrophic genus Azoarcus sensu lato and description of three new genera and new species, Azovibrio restrictus gen. nov., sp. nov., Azospira oryzae gen. nov., sp. nov. and Azonexus fungiphilus gen. nov., sp. nov. Int J Syst Evol Microbiol 50:649–659
Rikken G, Kroon A, van Ginkel C (1996) Transformation of (per)chlorate into chloride by a newly isolated bacterium: reduction and dismutation. Appl Microbiol Biotechnol 45:420–426
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574
Scanlon BR, Reedy RC, Jackson WA, Rao B (2008) Mobilization of naturally occurring perchlorate related to land-use change in the southern high plains, Texas. Environ Sci Technol 42:8648–8653
Shea C, Clauwaert P, Verstraete W, Nerenberg R (2008) Adapting a denitrifying biocathode for perchlorate reduction. Water Sci Technol 58:1941–1946
Shrout JD, Scheetz TE, Casavant TL, Parkin GF (2005) Isolation and characterization of autotrophic, hydrogen-utilizing, perchlorate-reducing bacteria. Appl Microbiol Biotechnol 67:261–268
Tanaka K, Nakamura K, Mikami E (1990) Fermentation of maleate by a gram-negative strictly anaerobic non-spore-former, Propionivibrio dicarboxylicus gen. nov., sp. nov.*. Arch Microbiol 154:323–328
Thrash JC, Coates JD (2008) Review: direct and indirect electrical stimulation of microbial metabolism. Environ Sci Technol 42:3921–3931
Thrash JC, VanTrump JI, Weber KA, Miller E, Achenbach LA, Coates JD (2007) Electrochemical stimulation of microbial perchlorate reduction. Environ Sci Technol 41:1740–1746
Wallace W, Ward T, Breen A, Attaway H (1996) Identification of an anaerobic bacterium which reduces perchlorate and chlorate as Wolinella succinogenes. J Ind Microbiol 16:68–72
Weber KA, Hedrick DB, Peacock AD, Thrash JC, White DC, Achenbach LA, Coates JD (2009) Physiological and taxonomic description of the novl autotrophic, metal oxidizing bacterium, Pseudogulbenkiania sp. strain 2002. Appl Microbiol Biotechnol 83:555–656
Wolff J (1998) Perchlorate and the thyroid gland. Pharmacol Rev 50:89–105
Wolterink A, Kim S, Muusse M, Kim IS, Roholl PJM, van Ginkel CG, Stams AJM, Kengen SWM (2005) Dechloromonas hortensis sp. nov. and strain ASK-1, two novel (per)chlorate-reducing bacteria, and taxonomic description of strain GR-1. Int J Syst Evol Microbiol 55:2063–2068
Wrighton KC, Agbo P, Warnecke F, Weber KA, Brodie EL, DeSantis TZ, Hugenholtz P, Anderson GL, Coates JD (2008) A novel ecological role of the Firmicutes identified in thermophilic microbial fuel cells. ISME J 2:1146–1156
Zhang HS, Bruns MA, Logan BE (2002) Chemolithoautotrophic perchlorate reduction by a novel hydrogen-oxidizing bacterium. Environ Microbiol 4:570–576
Acknowledgments
Support for this research to JDC was from grant DACA72-00-C-0016 from the US Department of Defense SERDP program.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Thrash, J.C., Pollock, J., Torok, T. et al. Description of the novel perchlorate-reducing bacteria Dechlorobacter hydrogenophilus gen. nov., sp. nov. and Propionivibrio militaris, sp. nov.. Appl Microbiol Biotechnol 86, 335–343 (2010). https://doi.org/10.1007/s00253-009-2336-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-009-2336-6