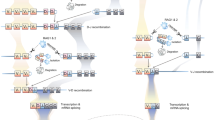
Abstract
Over 99% of ray-finned fishes (Actinopterygii) are teleosts, a clade that comprises half of all living vertebrate species that have diversified across virtually all fresh and saltwater ecosystems. This ecological breadth raises the question of how the immunogenetic diversity required to persist under heterogeneous pathogen pressures evolved. The teleost genome duplication (TGD) has been hypothesized as the evolutionary event that provided the substrate for rapid genomic evolution and innovation. However, studies of putative teleost-specific innate immune receptors have been largely limited to comparisons either among teleosts or between teleosts and distantly related vertebrate clades such as tetrapods. Here we describe and characterize the receptor diversity of two clustered innate immune gene families in the teleost sister lineage: Holostei (bowfin and gars). Using genomic and transcriptomic data, we provide a detailed investigation of the phylogenetic history and conserved synteny of gene clusters encoding diverse immunoglobulin domain-containing proteins (DICPs) and novel immune-type receptors (NITRs). These data demonstrate an ancient linkage of DICPs to the major histocompatibility complex (MHC) and reveal an evolutionary origin of NITR variable-joining (VJ) exons that predate the TGD by at least 50 million years. Further characterizing the receptor diversity of Holostean DICPs and NITRs illuminates a sequence diversity that rivals the diversity of these innate immune receptor families in many teleosts. Taken together, our findings provide important historical context for the evolution of these gene families that challenge prevailing expectations concerning the consequences of the TGD during actinopterygiian evolution.











Similar content being viewed by others
Data availability
All data used in this study are publicly available on NCBI, Ensembl, or the PhyloFish database (http://phylofish.sigenae.org/). Publicly available bowfin sequences accessed during this study are also provided as full length fasta formatted sequences and partitioned sequences linked to the delimitations of domains (e.g., NITR-I, NITR-V) in the Supplementary Materials.
References
Abi Rached L, McDermott MF, Pontarotti P (1999) The MHC big bang. Immunol Rev 167:33–44
Alfaro ME, Santini F, Brock C, Alamillo H, Dornburg A, Rabosky DL, Carnevale G, Harmon LJ (2009) Nine exceptional radiations plus high turnover explain species diversity in jawed vertebrates. Proc Natl Acad Sci U S A 106:13410–13414
Almagro Armenteros JJ, Tsirigos KD, Sønderby CK, Petersen TN, Winther O, Brunak S, von Heijne G, Nielsen H (2019) SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat Biotechnol 37:420–423
Aoki T, Hikima J-I, Hwang SD, Jung TS (2013) Innate immunity of finfish: primordial conservation and function of viral RNA sensors in teleosts. Fish Shellfish Immunol 35:1689–1702
Berv JS, Field DJ (2018) Genomic signature of an avian Lilliput effect across the K-Pg Extinction. Syst Biol 67:1–13
Betancur-R R, Wiley EO, Arratia G et al (2017) Phylogenetic classification of bony fishes. BMC Evol Biol 17:162
Boudinot P, Zou J, Ota T et al (2014) A tetrapod-like repertoire of innate immune receptors and effectors for coelacanths. J Exp Zool B Mol Dev Evol 322:415–437
Braasch I, Gehrke AR, Smith JJ et al (2016) The spotted gar genome illuminates vertebrate evolution and facilitates human-teleost comparisons. Nat Genet 48:427–437
Braasch I, Postlethwait JH (2012) Polyploidy in fish and the teleost genome duplication. In: Soltis P, Soltis D (eds) Polyploidy and genome evolution. Springer, Berlin, Heidelberg, pp 341–383
Brawand D, Wagner CE, Li YI et al (2014) The genomic substrate for adaptive radiation in African cichlid fish. Nature 513:375–381
Burns MD, Sidlauskas BL (2019) Ancient and contingent body shape diversification in a hyperdiverse continental fish radiation. Evolution 73:569–587
Cannon JP, Haire RN, Magis AT, Eason DD, Winfrey KN, Hernandez Prada JA, Bailey KM, Jakoncic J, Litman GW, Ostrov DA (2008) A bony fish immunological receptor of the NITR multigene family mediates allogeneic recognition. Immunity 29(2):228–237
Carmona SJ, Teichmann SA, Ferreira L et al (2017) Single-cell transcriptome analysis of fish immune cells provides insight into the evolution of vertebrate immune cell types. Genome Res 27:451–461
Conant GC (2020) The lasting after-effects of an ancient polyploidy on the genomes of teleosts. PLoS One 15:e0231356
Daane JM, Dornburg A, Smits P et al (2019) Historical contingency shapes adaptive radiation in Antarctic fishes. Nat Ecol Evol 3:1102–1109
Davis AM, Unmack PJ, Pusey BJ et al (2012) Marine-freshwater transitions are associated with the evolution of dietary diversification in terapontid grunters (Teleostei: Terapontidae). J Evol Biol 25:1163–1179
Davis MP, Holcroft NI, Wiley EO et al (2014) Species-specific bioluminescence facilitates speciation in the deep sea. Mar Biol 161:1139–1148
Desai S, Heffelfinger AK, Orcutt TM et al (2008) The medaka novel immune-type receptor (NITR) gene clusters reveal an extraordinary degree of divergence in variable domains. BMC Evol Biol 8:177
Dirscherl H, McConnell SC, Yoder JA, de Jong JLO (2014) The MHC class I genes of zebrafish. Dev Comp Immunol 46:11–23
Dornburg A, Federman S, Lamb AD et al (2017) Cradles and museums of Antarctic teleost biodiversity. Nat Ecol Evol 1:1379–1384
Dornburg A, Sidlauskas B, Santini F et al (2011) The influence of an innovative locomotor strategy on the phenotypic diversification of triggerfish (family: Balistidae). Evolution 65:1912–1926
Dornburg A, Townsend JP, Friedman M, Near TJ (2014) Phylogenetic informativeness reconciles ray-finned fish molecular divergence times. BMC Evol Biol 14:169
Fabbri M, Mongiardino Koch N, Pritchard AC et al (2017) The skull roof tracks the brain during the evolution and development of reptiles including birds. Nat Ecol Evol 1:1543–1550
Fernández R, Gabaldón T (2020) Gene gain and loss across the metazoan tree of life. Nat Ecol Evol 4:524–533
Ferraresso S, Kuhl H, Milan M et al (2009) Identification and characterisation of a novel immune-type receptor (NITR) gene cluster in the European sea bass, Dicentrarchus labrax, reveals recurrent gene expansion and diversification by positive selection. Immunogenetics 61:773–788
Friedman M, Keck BP, Dornburg A et al (2013) Molecular and fossil evidence place the origin of cichlid fishes long after Gondwanan rifting. Proc Biol Sci 280:20131733
Friedman ST, Martinez CM, Price SA, Wainwright PC (2019) The influence of size on body shape diversification across Indo-Pacific shore fishes*. Evolution 73:1873–1884
Gajdzik L, Aguilar-Medrano R, Frédérich B (2019) Diversification and functional evolution of reef fish feeding guilds. Ecol Lett 22:572–582
Gao F-X, Lu W-J, Wang Y et al (2018) Differential expression and functional diversification of diverse immunoglobulin domain-containing protein (DICP) family in three gynogenetic clones of gibel carp. Dev Comp Immunol 84:396–407
Glasauer SMK, Neuhauss SCF (2014) Whole-genome duplication in teleost fishes and its evolutionary consequences. Mol Genet Genomics 289:1045–1060
Grande L (2010) An empirical synthetic pattern study of gars (Lepisosteiformes) and closely related species, based mostly on skeletal anatomy: the resurrection of Holostei. Copeia 2010(2A):iii–871
Haire RN, Cannon JP, O’Driscoll ML et al (2012) Genomic and functional characterization of the diverse immunoglobulin domain-containing protein (DICP) family. Genomics 99:282–291
Hoang DT, Chernomor O, von Haeseler A et al (2018) UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol 35:518–522
Höhne C, Prokopov D, Kuhl H, Du K, Klopp C, Wuertz S, Trifonov V, Stöck M (2021) The immune system of sturgeons and paddlefish (Acipenseriformes): a review with new data from a chromosome-scale sturgeon genome. Rev Aquac 13(3):1709–1729
Holt C, Yandell M (2011) MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics 12:491
Hughes LC, Ortí G, Huang Y et al (2018) Comprehensive phylogeny of ray-finned fishes (Actinopterygii) based on transcriptomic and genomic data. Proc Natl Acad Sci U S A 115:6249–6254
Iglesias TL, Dornburg A, Warren DL et al (2018) Eyes wide shut: the impact of dim-light vision on neural investment in marine teleosts. J Evol Biol 31:1082–1092
Inoue J, Sato Y, Sinclair R et al (2015) Rapid genome reshaping by multiple-gene loss after whole-genome duplication in teleost fish suggested by mathematical modeling. Proc Natl Acad Sci U S A 112:14918–14923
Jones DT, Taylor WR, Thornton JM (1992) The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8:275–282
Kalyaanamoorthy S, Minh BQ, Wong TKF et al (2017) ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods 14:587–589
Kapustin Y, Souvorov A, Tatusova T, Lipman D (2008) Splign: algorithms for computing spliced alignments with identification of paralogs. Biol Direct 3:20
Kasahara M (1999) The chromosomal duplication model of the major histocompatibility complex. Immunol Rev 167:17–32
Kasahara M, Flajnik MF (2019) Origin and evolution of the specialized forms of proteasomes involved in antigen presentation. Immunogenetics 71:251–261
Katoh K, Standley DM (2014) MAFFT: iterative refinement and additional methods. Methods Mol Biol 1079:131–146
Langevin C, Aleksejeva E, Passoni G et al (2013) The antiviral innate immune response in fish: evolution and conservation of the IFN system. J Mol Biol 425:4904–4920
Lefranc M-P, Giudicelli V, Duroux P et al (2015) IMGT®, the international ImMunoGeneTics information system® 25 years on. Nucleic Acids Res 43:D413–D422
Letunic I, Bork P (2018) 20 years of the SMART protein domain annotation resource. Nucleic Acids Res 46:D493–D496
Litman GW, Hawke NA, Yoder JA (2001) Novel immune-type receptor genes. Immunol Rev 181:250–259
Litman GW, Yoder JA, Cannon JP, Haire RN (2003) Novel immune-type receptor genes and the origins of adaptive and innate immune recognition. Integr Comp Biol 43:331–337
Minh BQ, Nguyen MAT, von Haeseler A (2013) Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol 30:1188–1195
Montgomery BC, Cortes HD, Mewes-Ares J et al (2011) Teleost IgSF immunoregulatory receptors. Dev Comp Immunol 35:1223–1237
Nakamura T, Yamada KD, Tomii K, Katoh K (2018) Parallelization of MAFFT for large-scale multiple sequence alignments. Bioinformatics 34:2490–2492
Nakatani M, Miya M, Mabuchi K et al (2011) Evolutionary history of Otophysi (Teleostei), a major clade of the modern freshwater fishes: Pangaean origin and Mesozoic radiation. BMC Evol Biol 11:177
Near TJ, Dornburg A, Eytan RI et al (2013) Phylogeny and tempo of diversification in the superradiation of spiny-rayed fishes. Proc Natl Acad Sci U S A 110:12738–12743
Near TJ, Dornburg A, Kuhn KL et al (2012a) Ancient climate change, antifreeze, and the evolutionary diversification of Antarctic fishes. Proc Natl Acad Sci U S A 109:3434–3439
Near TJ, Eytan RI, Dornburg A et al (2012b) Resolution of ray-finned fish phylogeny and timing of diversification. Proc Natl Acad Sci U S A 109:13698–13703
Near TJ, Kim D (2021) Phylogeny and time scale of diversification in the fossil-rich sunfishes and black basses (Teleostei: Percomorpha: Centrarchidae). Mol Phylogenet Evol 161:107156
Nei M, Rooney AP (2005) Concerted and birth-and-death evolution of multigene families. Annu Rev Genet 39:121–152
Nesbitt SJ (2003) Arizonasaurus and its implications for archosaur divergence. Proc Biol Sci 270(Suppl 2):S234–S237
Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ (2015) IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274
Pasquier J, Cabau C, Nguyen T et al (2016) Gene evolution and gene expression after whole genome duplication in fish: the PhyloFish database. BMC Genomics 17:368
Pietretti D, Wiegertjes GF (2014) Ligand specificities of Toll-like receptors in fish: indications from infection studies. Dev Comp Immunol 43:205–222
Price SA, Schmitz L, Oufiero CE et al (2014) Two waves of colonization straddling the K-Pg boundary formed the modern reef fish fauna. Proc Biol Sci 281:20140321
Prum RO, Berv JS, Dornburg A et al (2015) A comprehensive phylogeny of birds (Aves) using targeted next-generation DNA sequencing. Nature 526:569–573
Quang LS, Gascuel O, Lartillot N (2008) Empirical profile mixture models for phylogenetic reconstruction. Bioinformatics 24:2317–2323
Rebl A, Goldammer T, Seyfert H-M (2010) Toll-like receptor signaling in bony fish. Vet Immunol Immunopathol 134:139–150
Rodríguez-Nunez I, Wcisel DJ, Litman GW, Yoder JA (2014) Multigene families of immunoglobulin domain-containing innate immune receptors in zebrafish: deciphering the differences. Dev Comp Immunol 46:24–34
Rodriguez-Nunez I, Wcisel DJ, Litman RT et al (2016) The identification of additional zebrafish DICP genes reveals haplotype variation and linkage to MHC class I genes. Immunogenetics 68:295–312
Salzburger W (2018) Understanding explosive diversification through cichlid fish genomics. Nat Rev Genet 19:705–717
Seehausen O (2006) African cichlid fish: a model system in adaptive radiation research. Proceedings of the Royal Society B: Biological Sciences 273:1987–1998
Shah RN, Rodriguez-Nunez I, Eason DD et al (2012) Development and characterization of anti-nitr9 antibodies. Adv Hematol 2012:596925
Sibert E, Friedman M, Hull P et al (2018) Two pulses of morphological diversification in Pacific pelagic fishes following the Cretaceous-Palaeogene mass extinction. Proceedings of the Royal Society B: Biological Sciences 285:20181194
Sievers F, Higgins DG (2018) Clustal Omega for making accurate alignments of many protein sequences. Protein Sci 27:135–145
Siqueira AC, Bellwood DR, Cowman PF (2019) Historical biogeography of herbivorous coral reef fishes: the formation of an Atlantic fauna. J Biogeo
Smithwick FM, Stubbs TL (2018) Phanerozoic survivors: Actinopterygian evolution through the Permo-Triassic and Triassic-Jurassic mass extinction events. Evolution 72:348–362
Soubrier J, Steel M, Lee MSY et al (2012) The influence of rate heterogeneity among sites on the time dependence of molecular rates. Mol Biol Evol 29:3345–3358
Strong SJ, Mueller MG, Litman RT et al (1999) A novel multigene family encodes diversified variable regions. Proc Natl Acad Sci U S A 96:15080–15085
Takezaki N (2018) Global rate variation in bony vertebrates. Genome Biol Evol 10:1803–1815
Thompson AW, Hawkins MB, Parey E et al (2021) The bowfin genome illuminates the developmental evolution of ray-finned fishes. Nat Genet. https://doi.org/10.1038/s41588-021-00914-y
Traver D, Yoder JA (2020) Chapter 19—Immunology. In: Cartner SC, Eisen JS, Farmer SC, et al. (eds) The zebrafish in biomedical research. Acad Press, pp 191–216
Tukwasibwe S, Nakimuli A, Traherne J et al (2020) Variations in killer-cell immunoglobulin-like receptor and human leukocyte antigen genes and immunity to malaria. Cell Mol Immunol 17:799–806
Uhrberg M, Parham P, Wernet P (2002) Definition of gene content for nine common group B haplotypes of the Caucasoid population: KIR haplotypes contain between seven and eleven KIR genes. Immunogenetics 54:221–229
Vilches C, Parham P (2002) KIR: diverse, rapidly evolving receptors of innate and adaptive immunity. Annu Rev Immunol 20:217–251
Wang Y, Tang H, Debarry JD et al (2012) MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res 40:e49
Wcisel DJ, Howard JT 3rd, Yoder JA, Dornburg A (2020) Transcriptome Ortholog Alignment Sequence Tools (TOAST) for phylogenomic dataset assembly. BMC Evol Biol 20:41
Wcisel DJ, Ota T, Litman GW, Yoder JA (2017) Spotted gar and the evolution of innate immune receptors. J Exp Zool B Mol Dev Evol 328(7):666–684
Wcisel DJ, Yoder JA (2016) The confounding complexity of innate immune receptors within and between teleost species. Fish Shellfish Immunol 53:24–34
Wei S, Zhou J-M, Chen X et al (2007) The zebrafish activating immune receptor Nitr9 signals via Dap12. Immunogenetics 59:813–821
Whelan S, Goldman N (2001) A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol 18:691–699
Wickham H (2011) ggplot2. Wiley Interdisciplinary Reviews: Computational Statistics 3:180–185
Williams AF, Barclay AN (1988) The immunoglobulin superfamily—domains for cell surface recognition. Annu Rev Immunol 6:381–405
Yamaguchi T, Dijkstra JM (2019) Major histocompatibility complex (MHC) genes and disease resistance in fish. Cells 8:378. https://doi.org/10.3390/cells8040378
Yamanoue Y, Miya M, Doi H et al (2011) Multiple invasions into freshwater by pufferfishes (teleostei: tetraodontidae): a mitogenomic perspective. PLoS One 6:e17410
Yang Z (1994) Maximum likelihood phylogenetic estimation from DNA sequences with variable rates over sites: approximate methods. J Mol Evol 39:306–314
Yoder JA (2009) Form, function and phylogenetics of NITRs in bony fish. Dev Comp Immunol 33:135–144
Yoder JA, Cannon JP, Litman RT et al (2008) Evidence for a transposition event in a second NITR gene cluster in zebrafish. Immunogenetics 60:257–265
Yoder JA, Litman GW (2011) The phylogenetic origins of natural killer receptors and recognition: relationships, possibilities, and realities. Immunogenetics 63:123–141
Yoder JA, Litman RT, Mueller MG et al (2004) Resolution of the novel immune-type receptor gene cluster in zebrafish. Proc Natl Acad Sci USA 101:15706–15711
Yoder JA, Turner PM, Wright PD et al (2010) Developmental and tissue-specific expression of NITRs. Immunogenetics 62:117–122
Acknowledgements
We thank Thomas Near (Yale University) for helpful discussions about bowfin biology and the consequences of the teleost genome duplication and Madhusudhan Gundappa (@fish_lines) for illustrations of bowfin and spotted gar.
Funding
This research was supported, in part, by grants from the National Science Foundation (IOS-1755242 to AD and IOS-1755330 to JAY), a grant from the Triangle Center for Evolutionary Medicine (TriCEM) to AD and JAY, and funding from the National Institutes of Health (R01OD011116 to IB).
Author information
Authors and Affiliations
Contributions
AD and JAY conceived of and designed the study. EF, KZ, AWT, IB, LRA, TO, DJW, and AWT assembled sequence data. AD, JAY, TO, EF, LRA, KZ, AWT, IB, and DJW analyzed the data. AD and JAY wrote the first draft of the manuscript. All authors contributed to the writing of the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Disclaimer
The funding bodies played no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Dornburg, A., Wcisel, D.J., Zapfe, K. et al. Holosteans contextualize the role of the teleost genome duplication in promoting the rise of evolutionary novelties in the ray-finned fish innate immune system. Immunogenetics 73, 479–497 (2021). https://doi.org/10.1007/s00251-021-01225-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00251-021-01225-6