Abstract
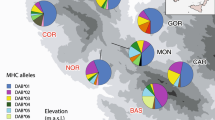
Many fishes express high levels of intraspecific variability, often linked to resource partitioning. Several studies show that a species’ evolutionary trajectory of adaptive divergence can undergo reversals caused by changes in its environment. Such a reversal in neutral genetic and morphological variation among lake trout Salvelinus namaycush ecomorphs appears to be underway in Lake Superior. However, a water depth gradient in neutral genetic divergence was found to be associated with intraspecific diversity in the lake. To investigate patterns of adaptive immunogenetic variation among lake trout ecomorphs, we used Illumina high-throughput sequencing. The population’s genetic structure of the major histocompatibility complex (MHC Class IIβ exon 2) and 18 microsatellite loci were compared to disentangle neutral and selective processes at a small geographic scale. Both MHC and microsatellite variation were partitioned more by water depth stratum than by ecomorph. Several metrics showed strong clustering by water depth in MHC alleles, but not microsatellites. We report a 75% increase in the number of MHC alleles shared between the predominant shallow and deep water ecomorphs since a previous lake trout MHC study at the same locale (c. 1990s data). This result is consistent with the reverse speciation hypothesis, although adaptive MHC polymorphisms persist along an ecological gradient. Finally, results suggested that the lake trout have multiple copies of the MHC II locus consistent with a historic genomic duplication event. Our findings indicated that conservation approaches for this species could focus on managing various ecological habitats by depth, in addition to regulating the fisheries specific to ecomorphs.





Similar content being viewed by others
References
Abbott R, Albach D, Ansell S, Arntzen JW, Baird SJE et al (2013) Hybridization and speciation. J Evol Biol 26:229–246. doi:10.1111/j.1420-9101.2012.02599.x
Aeschlimann PB, Häberli MA, Reusch TBH, Boehm T, Milinski M (2003) Female sticklebacks Gasterosteus aculeatus use self-reference to optimize MHC allele number during mate selection. Behav Ecol Sociobiol 54:119–126
Aguilar A, Garza JC (2006) A comparison of variability and population structure for major histocompatibility complex and microsatellite loci in California coastal steelhead (Oncorhynchus mykiss Walbaum). Mol Ecol 15:923–937
Allendorf FW, Thorgaard GH (1984) Tetraploidy and evolution of salmonid fishes: evolutionary genetics of fishes. Plenum Press, New York
Allendorf FW, Berry O, Ryman N (2014) So long to genetic diversity, and thanks for all the fish. Mol Ecol 23:23–25
Angers B, Bernatchez L, Angers A, Desgroseillers L (1995) Specific microsatellite loci for brook charr (Salvelinus fontinalis) reveal strong population subdivision on a microgeographic scale. J Fish Biol 47:177–185
Austin JA, Allen J (2011) Sensitivity of summer Lake Superior thermal structure to meteorological forcing. Limnol Oceanogr 56:1141–1154
Baillie SM, Muir AM, Scribner K, Bentzen P, Krueger CC (2016a) Loss of genetic diversity and reduction of genetic distance among lake trout Salvelinus namaycush ecomorphs, Lake Superior 1959 to 2013. J Great Lakes Res 42:204–216. doi:10.1016/j.jglr.2016.02.001
Baillie SM, Muir AM, Krueger CC, Scribner K, Bentzen P (2016b) Genetic and phenotypic variation along an ecological gradient in lake trout Salvelinus namaycush. BMC Evol Biol 16:219. doi:10.1186/s12862-016-0788-8
Bernatchez L, Landry C (2003) MHC studies in nonmodel vertebrates: what have we learned about natural selection in 15 years? J Evolution Biol 16:363–377
Borghans JAM, Beltman JB, De Boer RJ (2004) MHC polymorphism under host-pathogen coevolution. Immunogenetics 55:732–739. doi:10.1007/s00251-003-0630-5
Bronte CR, Ebener MP, Schreiner DR, DeVault DS, Petzold MM, Jensen DA, Richards C, Lozano SJ (2003) Fish community change in Lake Superior, 1970–2000. Can J Fish Aquat Sci 60:1552–1574
Cammen K, Hoffmann JI, Knapp LA, Harwood J, Amos W (2011) Geographic variation of the major histocompatibility complex in Eastern Atlantic grey seals (Halichoerus grypus). Mol Biol 20:740–752
Carvajal-Rodriguez A, de Una-Alvarez J (2011) Assessing significance in high throughput experiments by sequential goodness of fit and q-value estimation. PLoS Genet 6:e24700
Chain FJJ, Feulner PGD, Panchal M, Eizaguirre C, Samonte IE et al (2014) Extensive copy-number variation of young genes across stickleback populations. PLoS Genet 10:e1004830. doi:10.1371/journal.pgen.1004830
Chavarie L, Howland KL, Tonn WM (2013) Sympatric polymorphism in lake trout: the coexistence of multiple shallow-water morphotypes in Great Bear Lake. Trans Am Fish Soc 142:814–823
Cheng Y, Stuart A, Morris K, Taylor R, Siddle H, Deakin J, Jones M, Amemiya CT, Belov K (2012) Antigen-presenting genes and genomic copy number variations in the Tasmanian devil MHC. BMC Genomics 13:87. doi:10.1186/1471-2164-13-87
R Core Team (2015) R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing. https://www.r-project.org
Croisetiere S, Tarte P, Bernatchez L, Belhumeur P (2008) Identification of MHC class IIβ resistance/susceptibility alleles to Aeromonas salmonicida in brook charr (Salvelinus fontinalis). Mol Immunol 45:3107–3116
De Leon LF, Raeymaekers JAM, Bermingham E, Podos J, Herrel A, Hendry AP (2011) Exploring possible human influences on the evolution of Darwin’s finches. Evolution 65:2258–2272. doi:10.1111/j.1558-5646.2011.01297.x
Delport W, Scheffler K, Seoighe C (2010) Frequent toggling between alternative amino acids is driven by selection in HIV-1. PLoS Genet. doi:10.1371/journal.ppat.1000242
Dionne M, Miller KM, Dodson JJ, Caron F, Bernatchez L (2007) Clinal variation in MHC diversity with temperature: evidence for the role of host-pathogen interaction on local adaptation in Atlantic salmon. Evolution 61:2154–2164
Dionne M, Miller KM, Dodson JJ, Bernatchez L (2009) MHC standing genetic variation and pathogen resistance in wild Atlantic salmon. Phil Trans Roy Soc B 364:1555–1565
Dorschner MO, Duris T, Bronte CR, Burnham Curtis MK, Phillips RB (2000) High levels of MHC class II allelic diversity in lake trout from Lake Superior. J Hered 91:359–363
Earl DA, von Holdt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361
Eizaguirre C, Baltazar-Soares M (2014) Evolutionary conservation—evaluating the adaptive potential of species. Evol Appl 7:963–967. doi:10.1111/eva.12227
Ejsmond MJ, Babik W, Radwan J (2010) MHC allele frequency distributions under parasite-driven selection: a simulation model. BMC Evol Biol 10:332
Ekblom R, Saether SA, Jacobsson P, Fiske P, Sahlman T, Grahn M, Kålås JA, Höglund J (2007) Spatial pattern of MHC class II variation in the great snipe (Gallinago media). Mol Ecol 16:1439–1451
Elphinstone MS, Hinten GN, Anderson MJ, Nock CJ (2003) An inexpensive and high-throughput procedure to extract and purify total genomic DNA for population studies. Mol Ecol Notes 3:317–320
Englbrecht C (2000) The impact of stocking on the genetic integrity of Arctic charr (Salvelinus) populations from the alpine region. Mol Ecol 11:1017–1027
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Excoffier L, Lischer HEL (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10:564–567
Falush D, Stephens M, Pritchard JK (2007) Inference of population structure using multilocus genotype data: dominant markers and null alleles. Mol Ecol Notes 7:574–578
Fraley C, Raftery AE (2009) MCLUST version 3 for R: normal mixuture modeling and model-based clustering. Technical report 504. University of Washington, Seattle https://www.stat.washington.edu/research/reports/2012/tr504.pdf
Garrigan D, Hedrick PW (2003) Perspective: detecting adaptive molecular polymorphism: lessons from the MHC. Evolution 57:1707–1722
Goyette P, Boucher G, Mallon D, Ellinghaus E, Jostins L et al (2015) High-density mapping of the MHC identifies a shared role for HLA-DRB1[ast]01:03 in inflammatory bowel diseases and heterozygous advantage in ulcerative colitis. Nat Genet 47:172–179. doi:10.1038/ng.3176
Great Lakes Fishery Commission (2011). Strategic vision of the Great Lakes fishery commission 2011–2020. Ann Arbor, Michigan. http://www.glfc.org/pubs/SpecialPubs/StrategicVision2012.pdf
Grimholt U, Tsukamoto K, Azuma T, Leong J, Koop BF, Dijkstra JM (2015) A comprehensive analysis of teleost MHC class I sequences. BMC Evol Biol 15:32. doi:10.1186/s12862-015-0309-1
Guinand B, Scribner KT, Page KS, Burnham-Curtis MK (2003) Genetic variation over space and time: analyses of extinct and remnant lake trout populations in the upper Great Lakes. Proc Roy Soc B 270:425–433
Guinand B, Page KS, Burnham-Curtis MK, Scribner KT (2012) Genetic signatures of historical bottlenecks in sympatric lake trout (Salvelinus namaycush) morphotypes in Lake Superior. Environ Biol Fish 95:323–334
Herdegen M, Babik W, Radwan J (2014) Selective pressures on MHC class II genes in the guppy (Poecilia reticulata) as inferred by hierarchical analysis of population structure. J Evol Biol 27:2347–2359. doi:10.1111/jeb.12476
Holeck KT, Mills EL, MacIsaac HJ, Dochoda MR, Colautti RI, Ricciardi A (2004) Bridging troubled waters: biological invasions, transoceanic shipping, and the Laurentian Great Lakes. Bioscience 54:919–929. doi:10.1641/0006-3568
Hubisz MJ, Falush D, Stephens M, Pritchard JK (2009) Inferring weak population structure with the assistance of sample group information. Mol Ecol Resour 9:1322–1332
Hughes AL, Yeager M (1998) Natural selection and the evolutionary history of major histocompatibility complex loci. Front Biosci 3:509–516
Kalinowski S (2004) Counting alleles with rarefaction: private alleles and hierarchical sampling designs. Conserv Genet 5:539–543
Kamath PL, Getz WM (2012) Unraveling the effects of selection and demography on immune gene variation in free-ranging plains zebra (Equus quagga) populations. PLoS Genet 7:e50971
King TL, Lubinski BA, Burnham-Curtis MK, Stott W, Morgan RP II (2012) Tools for the management and conservation of genetic diversity in brook trout (Salvelinus fontinalis): tri- and tetranucleotide microsatellite markers for the assessment of genetic diversity, phylogeography, and historical demographics. Mol Ecol Resour 4:539–543
Knafler GJ, Grueber CE, Sutton JT, Jamieson IG (2017) Differential patterns of diversity at microsatellite, MHC, and TLR loci in bottlenecked South Island saddleback populations. New Zeal J Ecol 41:98–106. doi:10.20417/nzjecol.41.8
Kosakovsky-Pond SL, Frost SWD, Muse SV (2005) HyPhy: hypothesis testing using phylogenies. Bioinformatics 21:676–679
Krueger CC, Ihssen PE (1995) Review of genetics of lake trout in the Great Lakes: history, molecular genetics, physiology, strain comparisons, and restoration management. J Great Lakes Res 21:348–363
Lamaze FC, Pavey SA, Normandeau E, Roy G, Garant D, Bernatchez L (2014) Neutral and selective processes shape MHC gene diversity and expression in stocked brook charr populations (Salvelinus fontinalis). Mol Ecol 23:1730–1748
Landry C, Bernatchez L (2001) Comparative analysis of population structure across environments and geographical scales at major histocompatibility complex and microsatellite loci in Atlantic salmon (Salmo salar). Mol Ecol 10:2525–2539
Larsen DP, Kaufmann P, Kincaid TM, Urquhart NS (2004) Detecting persistent changes in the habitat of salmon-bearing streams in the Pacific Northwest. Can J Fish Aquat Sci 61:283–291
Larson WA, Lisi PJ, Seeb JE, Seeb LW, Schindler DE (2016) Major histocompatibility complex diversity is positively associated with stream water temperatures in proximate populations of sockeye salmon. J Evol Biol 29:1846–1859. doi:10.1111/jeb.12926
Lee JH, Jeon JT (2008) Methods to detect and analyze copy number variations at the genome-wide and locus-specific levels. Cytogenet Genome Res 123:333–342. doi:10.1159/000184725
Lighten J, Van Oosterhout C, Paterson IG, McMullan M, Bentzen P (2014) Ultra-deep Illumina sequencing accurately identifies MHC class IIβ alleles and provides evidence for copy number variation in the guppy (Poecilia reticulata). Mol Ecol Resour 14:753–767
Loch TP, Faisal M (2015) Emerging flavobacterial infections in fish: a review. J Adv Res 6:283–300. doi:10.1016/j.jare.2014.10.009
Luo MF, Pan HJ, Liu ZJ, Li M (2012) Balancing selection and genetic drift at major histocompatibility complex class II genes in isolated populations of golden snub-nosed monkey (Rhinopithecus roxellana). BMC Evol Biol 12:207
McGowan CR, Davidson EA, Woram RA, Danzmann RG, Ferguson MM, Davidson WS (2004) Ten polymorphic microsatellite markers from Arctic charr (Salvelinus alpinus): linkage analysis and amplification in other salmonids. Animal Genet 35:479–481
Miller KM, Withler RE, Beacham TD (1997) Molecular evolution at MHC genes in two populations of Chinook salmon (Onchorhynchus tshawytscha). Mol Ecol 6:937–954
Miller KM, Kaukinen KH, Beacham TD, Withler RE (2001) Geographic heterogeneity in natural selection on an MHC locus in sockeye salmon. Genetica 111:237–257
Miller KM, Kaukinen KH, Schulze AD (2002) Expansion and contraction of major histocompatibility complex genes: a teleostean example. Immunogenetics 53:941–963
Miller KM, Li S, Ming TJ, Kaukinen KH, Schulze AD (2006) The salmonid MHC class I: more ancient loci uncovered. Immunogenetics 58:571–589
Muir AM, Bronte CR, Zimmerman MS, Quinlan HR, Glase JD, Krueger CC (2014) Ecomorphological diversity of lake trout at Isle Royale, Lake Superior. Trans Am Fish Soc 143:972–987
Muir AM, Hansen M, Bronte CR, Krueger C (2015) If Arctic charr Salvelinus alpinus is ‘the most diverse vertebrate’, what is the lake charr Salvelinus namaycush? Fish Fish 16:1–14
Murrell B, Wertheim JO, Moola S, Weighill T, Scheffler K, Kosakovsky-Pond SL (2012) Detecting individual sites subject to episodic diversifying selection. PLoS Genet. doi:10.1371/journal.pgen.1002764
Murrell B, Moola S, Mabona A, Weighill T, Sheward D, Kosakovsky-Pond SL, Scheffler K (2013) FUBAR: a fast, unconstrained bayesian approximation for inferring selection. Mol Biol Evol 30:1196–1205. doi:10.1093/molbev/mst030
Noakes MA, Reimer T, Phillips RB (2003) Genotypic characterization of an MHC class II locus in lake trout (Salvelinus namaycush) from Lake Superior by single-stranded conformational polymorphism analysis and reference strand-mediated conformational analysis. Mar Biotechnol 5:270–278
Nordmo R, Ramstad A (1999) Variables affecting the challenge pressure of Aeromonas salmonicida and Vibrio salmonicida in Atlantic salmon (Salmo salar L.) Aquaculture 171:1–12
Nowak MA, Tarczy-Hornoch K, Austyn JM (1992) The optimal number of major histocompatibility complex molecules in an individual. Proc Natl Acad Sci U S A 89:10896–10899
Ohno S (1970) Evolution by gene duplication. Springer-Verlag, New York. doi:10.1007/978-3-642-86659-3
Oliver MK, Lambin X, Cornulier T, Piertney SB (2009) Spatio-temporal variation in the strength and mode of selection acting on major histocompatibility complex diversity in water vole (Arvicola terrestris) metapopulations. Mol Ecol 18:80–92
Olsen JB, Bentzen P, Seeb JE (1998) Characterization of seven microsatellite loci derived from pink salmon. Mol Ecol 7:1087–1089
O'Reilly PT, Hamilton LC, McConnell SK, Wright JM (1996) Rapid analysis of genetic variation in Atlantic salmon (Salmo salar) by PCR multiplexing of dinucleotide and tetranucleotide microsatellites. Can J Fish Aquatic Sci 53:2292–2298
Page K, Scribner K, Burnham-Curtis M (2004) Genetic diversity of wild and hatchery lake trout populations: relevance for management and restoration in the Great Lakes. Trans Am Fish Soc 133:674–691
Pavey SA, Lamaze FC, Garant D, Bernatchez L (2011) Full length MHC IIβ exon 2 primers for salmonids: a new resource for next generation sequencing. Conserv Genet Resour 3:665–667
Peakall R, Smouse PE (2006) GenAlEx 6: genetic analysis in Excel—population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel—population genetic software for teaching and research-an update. Bioinformatics 28:2537–2539
Perreault-Payette A, Muir AM, Goetz F, Perrier C, Normandeau E, Sirois P, Bernatchez L (2017) Investigating the extent of parallelism in morphological and genomic divergence among lake trout ecotypes in Lake Superior. Mol Ecol. doi:10.1111/mec.14018
Pigliucci M, Murren CJ, Schlichting CD (2006) Phenotypic plasticity and evolution by genetic assimilation. J Exp Biol 209:2362–2367
Pinsky ML, Palumbi SR (2014) Meta-analysis reveals lower genetic diversity in overfished populations. Mol Ecol 23:29–39
Pritchard J, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Pruett C, Winker K (2008) The effects of sample size on population genetic diversity estimates in song sparrows Melospiza melodia. J Avian Biol 39:252–256
Reed DH, Frankham R (2003) Correlation between fitness and genetics diversity. Conserv Biol 17:230–237. doi:10.1046/j.1523-1739.2003.01236.x
Rico Y, Morris-Pocock J, Zigouris J, Nocera JJ, Kyle CJ (2015) Lack of spatial immunogenetic structure among wolverine (Gulo gulo) populations suggestive of broad scale balancing selection. PLoS Genet 10:e0140170
Rico Y, Ethier DM, Davy CM, Sayers J, Weir RD, Swanson BJ, Nocera JJ, Kyle CJ (2016) Spatial patterns of immunogenetic and neutral variation underscore the conservation value of small, isolated American badger populations. Evol Appl 9:1271–1284. doi:10.1111/eva.12410
Rohlf JF (2015) Thin Plate Spline (TPS) suite of programs. http://life.bio.sunysb.edu/morph/
Rollins MF, Vu NV, Spies IB, Kalinowski ST (2009) Twelve microsatellite loci for lake trout (Salvelinus namaycush). Permanent Genet Resour Note 9:871–873. doi:10.1111/j.1755-0998.2008.02403
Scheffler K, Martin DP, Seoighe C (2006) Robust inference of positive selection from recombining coding sequences. Bioinformatics 22:2493–2499. doi:10.1093/bioinformatics/btl427
Scribner KT, Gust JR, Fields RL (1996) Isolation and characterization of novel salmon microsatellite loci: cross-species amplification and population genetics applications. Can J Fish Aquat Sci 53:833–841
Scribner KT, Lowe WH, Landguth E, Luikart G, Infante DM, Whelan GE, Muhlfeld CC (2016) Applications of genetic data to improve management and conservation of river fishes and their habitats. Fisheries 41:178–188. doi:10.1080/03632415.2016.1150838
Shum BP, Guethlein L, Flodin LR, Adkison MA, Hedrick RP, Nehring RB, Stet RJM, Secombes C, Parham P (2001) Modes of salmonid MHC class I and II evolution differ from the primate paradigm. J Immunol 166:3297–3308
Spurgin LG, Richardson DS (2010) How pathogens drive genetic diversity: MHC, mechanisms and misunderstandings. Phil Trans Roy Soc London Ser B 277:979–988
Strand TM, Segelbacher G, Quintela M, Xiao L, Axelsson T, Höglund J (2012) Can balancing selection on MHC loci counteract genetic drift in small fragmented populations of black grouse? Ecol Evol 2:341–353
Stuglik MT, Radwan J, Babik W (2011) jMHC: software assistant for multilocus genotyping of gene families using next-generation amplicons sequencing. Mol Ecol Resour 11:739–742
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Tobler M, Plath M, Riesch R, Schlupp I, Grasse A et al (2014) Selection from parasites favours immunogenetic diversity but not divergence among locally adapted host populations. J Evol Biol 27:960–974
Van Oosterhout C, Hutchinson W, Wills D, Shipley P (2004) MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4:535–538
Van Oosterhout C, Joyce DA, Cummings SM, Blais J, Barson NJ et al (2006) Balancing selection, random genetic drift, and genetic variation at the major histocompatibility complex in two wild populations of guppies (Poecilia reticulata). Evolution 60:2562–2574
Volis S, Ormanbekova D, Yermekbayev K (2015) Role of phenotypic plasticity and population differentiation in adaptation to novel environmental conditions. Ecol Evol 5:3818–3829. doi:10.1002/ece3.1607
Williamson KS, Cordes JF, May BP (2002) Characterization of microsatellite loci in Chinook salmon (Oncorhynchus tshawytscha) and cross species amplification in other salmonids. Mol Ecol Notes 2:17–19
Woelfing B, Traulsen A, Milinski M, Boehm T (2009) Does intra-individual major histocompatibility complex diversity keep a golden mean? Phil Trans Roy Soc London Ser B 364:117–128
Zheng D, Mai K, Liu S, Cao L, Liufu Z, Xu W, Tan B, Zhang W (2004) Effect of temperature and salinity on virulence of Edwardsiella tarda to Japanese flounder, Paralichthys olivaceus (Temminck et Schlegel). Aquac Res 35:494–500
Zimmerman MS, Krueger CC (2009) An ecosystem perspective on re-establishing native deepwater fishes in the Laurentian Great Lakes. N Am J Fish Manag 29:1352–1371
Zimmerman MS, Krueger CC, Eshenroder RL (2006) Phenotypic diversity of lake trout in Great Slave Lake: differences in morphology, buoyancy, and habitat depth. Trans Am Fish Soc 135:1056–1067
Zimmerman MS, Krueger CC, Eshenroder RL (2007) Morphological and ecological differences between shallow- and deep-water lake trout in Lake Mistassini, Quebec. J Great Lakes Res 33:156–169
Acknowledgements
Two anonymous reviewers and the handling editor of Immunogenetics provided helpful comments on this manuscript. M. McBride, L. Anstey, and C. Angelidis (Marine Gene Probe lab, Dalhousie University) helped with microsatellite genotyping work at various stages of the project. We thank S. Sivertson, E. Strom, and B. Strom for their hospitality at Washington and Barnam Islands, adjacent to Isle Royale. J. Pyatskowit, C.R. Bronte, M.S. Zimmerman, H.R. Quinlan, and J.D. Glase provided cheerful and able assistance with the Isle Royale fieldwork. This research was funded by the Great Lakes Fishery Commission grant 2005_BEN_44001.
Author information
Authors and Affiliations
Contributions
S.M.B. wrote the article based on R.H.’s Honours Thesis work, which focused on MHC diversity among lake trout at Isle Royale, Lake Superior. MHC labwork and genotyping was performed by S.M.B. and R.H. Microsatellite labwork and genotyping was performed by S.M.B. at the latter stages of the project. S.M.B., P.B., C.C.K., and A.M.M. designed the study and managed the project. C.C.K. and A.M.M. obtained funding from the Great Lakes Fishery Commission and performed fieldwork to collect and process contemporary samples. All authors read, edited, and approved this version of the manuscript.
Corresponding author
Ethics declarations
Funding
This research was funded by the Great Lakes Fishery Commission grant 2005_BEN_44001.
Conflict of interest
The authors declare that they have no conflict of interest.
Informed consent
Informed consent was obtained from all individual participants included in the study. Additional informed consent was obtained from all individual participants for whom identifying information is included in this article.
Ethical approval
All applicable international, national, and/or institutional guidelines for the care and use of animals were followed. Sampling and handling of fish was in accordance with the guidelines for the care and use of fishes by the American Fisheries Society (http://fisheries.org/docs/wp/Guidelines-for-Use-of-Fishes.pdf).
Rights and permissions
About this article
Cite this article
Baillie, S.M., Hemstock, R.R., Muir, A.M. et al. Small-scale intraspecific patterns of adaptive immunogenetic polymorphisms and neutral variation in Lake Superior lake trout. Immunogenetics 70, 53–66 (2018). https://doi.org/10.1007/s00251-017-0996-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00251-017-0996-4