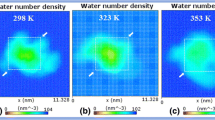
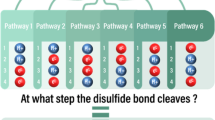
Abstract
Protein folding is a multi-micro second time scale event and involves many conformational transitions. Crucial conformational transitions responsible for biological functions of biomolecules are difficult to capture using current state-of-the-art molecular dynamics (MD) simulations. Protein folding, being a stochastic process, witnesses these transitions as rare events. Many new methodologies have been proposed for observing these rare events. In this work, a temperature-aided cascade MD is proposed as a technique for studying the conformational transitions. Folding studies for Engrailed homeodomain and Immunoglobulin domain B of protein A have been carried out. Using this methodology, the unfolded structures with RMSD of 20 Å were folded to a structure with RMSD of 2 Å. Three sets of cascade MD runs were carried out using implicit solvation, explicit solvation, and charge updation scheme. In the charge updation scheme, charges based on the conformation obtained are calculated and are updated in the topology file. In all the simulations, the structure of 2 Å was reached within a few nanoseconds using these methods. Umbrella sampling has been performed using snapshots from the temperature-aided cascade MD simulation trajectory to build an entire conformational transition pathway. The advantage of the method is that the possible pathways for a particular reaction can be explored within a short duration of simulation time and the disadvantage is that the knowledge of the start and end state is required. The charge updation scheme adds the polarization effects in the force fields. This improves the electrostatic interaction among the atoms, which may help the protein to fold faster.














Similar content being viewed by others
References
Anfinsen CB (1973) Principles that govern the folding of protein chains. Science 181:223–230
Berendsen HJC, Postma JPM, Vangunsteren WF, Dinola A, Haak JR (1984) Molecular dynamics with coupling to an external bath. J Chem Phys 81:3684
Case D, Darden T, Cheatham III TE, Simmerling C, Wang J, Duke R, Luo R, Walker R, Zhang W, Merz K (2012) AMBER 12. University of California, San Francisco
Caves LSD, Evanseck JD, Karplus M (1998) Locally accessible conformations of proteins: multiple molecular dynamics simulations of crambin. Protein Sci 7:649
Clarke ND, Kissinger CR, Desjarlais J, Gilliland GL, Pabo CO (1994) Structural studies of the engrailed homeodomain. Protein Sci 3:1779–1787
DeMarco ML, Alonso DOV, Daggett V (2004) Diffusing and colliding: the atomic level folding/unfolding pathway of a small helical protein. J Mol Biol 34:1109–1124
Dill KA, Chan HS (1997) From Levinthal to pathways to funnels. Nat Struct Biol 4(1):10–19 (Review)
Duan Y, Wang L, Kollman PA (1998) Pathways to a protein folding intermediate observed in a 1-microsecond simulation in aqueous solution. Proc Natl Acad Sci USA 95:9897–9902
Duan LL, Mei Y, Zhang D, Zhang QG, Zhang JZ (2010) Folding of a helix at room temperature is critically aided by electrostatic polarization of intraprotein hydrogen bonds. J Am Chem Soc 132(32):11159–11164
Gianni S, Guydosh NR, Khan F, Caldas TD, Mayor U, White GWN, DeMarco ML, Daggett V, Fresht A (2003) Unifying features in protein-folding mechanisms. Proc Natl Acad Sci 100:13286–13291
Götz AW, Williamson MJ, Xu D, Poole D, Le Grand S, Walker RC (2012) Microsecond molecular dynamics simulations with AMBER on GPUs. 1. Generalized born. J Chem Theory Comput 8:1542–1555
Gouda H, Torigoe H, Saito A, Sato M, Arata Y, Shimada I (1992) Three-dimensional solution structure of the B domain of staphylococcal protein A: comparisons of the solution and crystal structures. Bio-chemistry 31(40):9665–9672
Hamelberg D, Mongan J, McCammon J (2004) Accelerated molecular dynamics: a promising and efficient simulation method for biomolecules. J Chem Phys 120(24):11919–11929
Harada R, Kitao A (2013) Parallel Cascade Selection Molecular Dynamics (PaCS-MD) to generate conformational transition pathway. J Chem Phys 139(3):035103
Hawkins GD, Cramer CJ, Truhlar DG (1996) Parametrized models of aqueous free energies of solvation based on pair wise descreening of solute atomic charges from a dielectric medium. J Phys Chem 100:19824–19839
Huang F, Settanni G, Fresht A (2008) Fluorescence resonance energy transfer analysis of the folding pathway of engrailed homeodomain. Protein Eng Des Sel 21:131–146
Izrailev S, Crofts AR, Berry EA, Schulten K (1999) Steered molecular dynamics simulation of the Rieske subunit motion in the cytochrome bc(1) complex. Biophys J 77(4):1753–1768
Jani V, Sonavane U, Joshi R (2011) Microsecond scale replica exchange molecular dynamic simulation of villin headpiece: an insight into the folding landscape. J Biomol Struct Dyn 28(6):845–860
Jani V, Sonavane UB, Joshi R (2014) REMD and umbrella sampling simulations to probe the energy barrier of the folding pathways of engrailed homeodomain. J Mol Model 20(6):2283
Ji C, Mei Y (2014) Some practical approaches to treating electrostatic polarization of proteins. Acc Chem Res 47(9):2795–2803
Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML (1983) Comparison of simple potential functions for simulating liquid water. J Chem Phys 79:926
Koulgi S, Sonavane U, Joshi R (2010) Insights into the folding pathway of the engrailed homeodomain protein using replica exchange molecular dynamics simulations. J Mol Graph Model 29(3):481–491
Kumar S, Rosenberg JM, Bouzida D, Sendsen RH, Kollman PA (1992) The weighted histogram analysis method for free-energy calculations on biomolecules I. The method. J Comp Chem 13:1011–1021
Kuwajima K, Schmid KFX (1984) Experimental studies of folding kinetics and structural dynamics of small proteins. Adv Biophys 18:43–74 (Review)
Laio A, Parrinello M (2002) Escaping free-energy minima. Proc Natl Acad Sci USA 99:12562–12566
Lazim R, Wei C, Sun T, Zhang D (2013) Ab initio folding of extended α-helix: a theoretical study about the role of electrostatic polarization in the folding of helical structures. Proteins 81(9):1610–1620
Lei H, Wu C, Wang ZX, Zhou Y, Duan Y (2008) Folding processes of the B domain of protein A to the native state observed in all-atom ab initio folding simulations. J Chem Phys 128(23):235105
Li D, Haijun Y, Han L, Huo S (2008) Predicting the folding pathway of engrailed homeodomain with a probabilistic roadmap enhanced reaction-path algorithm. Biophys J 94:1622–1629
Linhananta A, Zhou HY, Zhou YQ (2002) The dual role of a loop with low loop contact distance in folding and domain swapping. Protein Sci 11:1695
Maaß A, Tekin ED, Schüller A, Palazoglu A, Reith D, Faller R (2010) Folding and unfolding characteristics of short beta strand peptides under different environmental conditions and starting configurations. Biochim Biophys Acta 1804:2003–2015
Marrink SJ, Risselada HJ, Yefimov S, Tieleman DP, de Vries AH (2007) The MARTINI force field: coarse-grained model for biomolecular simulations. J Phys Chem B 111(27):7812–7824
Mayor U, Johnson CM, Daggett V, Fersht AR (2000) Protein folding and unfolding in microseconds to nanoseconds by experiment and simulation. Proc Natl Acad Sci 97(25):13518–13522
McCully ME, Beck DA, Fersht A, Daggett AV (2010) Refolding the engrailed homeodomain: structural basis for the accumulation of a folding intermediate. Biophys J 99:1628–1636
MOPAC2012, James J. P. Stewart (2012) Stewart computational chemistry, Colorado Springs, CO, USA, http://OpenMOPAC.net
Myers JK, Oas TG (2001) Preorganized secondary structure as an important determinant of fast protein folding. Nat Struct Biol 8:552–558
Nakano M, Watanabe H, Rothstein SM, Tanaka S (2010) Comparative characterization of short monomeric polyglutamine peptides by replica exchange molecular dynamics simulation. J Phys Chem B 114:10234
Onuchic JN, Luthey-Schulten Z, Wolynes PG (1997) Theory of protein folding: the energy landscape perspective. Annu Rev Phys Chem 48:545–600
Peng C, Zhang L, Head-Gordon T (2010) Instantaneous normal modes as an unforced reaction coordinate for protein conformational transitions. Biophys J 98:2356
Radford SE, Dobson CM (1999) From computer simulations to human disease: emerging themes in protein folding. Cell 97:291–298
Religa TL, Johnson CM, Dung MV, Brewer SH, Dyer RB, Fresht A (2007) The helix–turn–helix as an ultrafast independently folding domain: the pathway of folding of engrailed homeodomain. Proc Natl Acad Sci 104:9272–9277
Shaw DE, Dror RO, Salmon JK, Grossman JP, Mackenzie KM, Bank JA, Cliff Y, Deneroff MM, Brannon B, Bowers KJ, Edmond C, Eastwood MP, Ierardi DJ, Klepeis JL, Kuskin JS, Larson RH, Kresten L-L, Paul M, Moraes MA, Stefano P, Yibing S, Brian T (2009) Millisecond-scale molecular dynamics simulations on Anton (SC09). ACM, New York
Suenaga A (2003) Replica-exchange molecular dynamics simulations for a small-sized protein folding with implicit solvent. THEOCHEM 634:235–241
Sugita Y, Okamoto Y (1999) Replica-exchange molecular dynamics method for protein folding. Chem Phys Lett 314(141–151):30
Thomas PJ, Qu BH, Pedersen PL (1995) Defective protein folding as a basis of human disease. Trends Biochem Sci 20:456–459
Torrie GM, Valleau JP (1977) Nonphysical sampling distributions in Monte Carlo free-energy estimation—umbrella sampling. J Comp Phys 23(187–199):35
Voelz VA, Bowman GR, Beauchamp K, Pande VS (2010) Molecular simulation of ab initio protein folding for a millisecond folder NTL9(1–39). J Am Chem Soc 132:1526–1528
Yang L, Tan CH, Hsieh MJ, Wang J, Duan Y, Cieplak P, Caldwell J, Luo R, Kollman PA (2006) New-generation amber united-atom force field. J Phys Chem B 110:13166–13176
Zagrovic B, Snow CD, Shirts MR, Pande VS (2002) Simulation of folding of a small alpha-helical protein in atomistic detail using worldwide-distributed computing. J Mol Biol 323:927–937
Zgarbová M, Otyepka M, Sponer J, Mladek A, Banas P, Cheatham III TE, Jurecka P (2011) Refinement of the Cornell et al. nucleic acids force field based on reference quantum chemical calculations of glycosidic torsion profiles. J Chem Theory Comp 7:2886–2902
Zhou YQ, Linhananta A (2002) Thermodynamics of an all-atom off-lattice model of the fragment B of Staphylococcal protein A: implication for the origin of the cooperativity of protein folding. J Phys Chem B 106:1481
Acknowledgments
The authors gratefully acknowledge the Department of Electronics and Information Technology (DeitY), Government of India, New Delhi, for providing financial support. This work was performed using the “Bioinformatics Resources and Applications Facility (BRAF)” and “National PARAM Supercomputing Facility (NPSF)” at C-DAC, Pune.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
249_2016_1115_MOESM1_ESM.docx
Supplementary Table 1 denotes the numbering of the atoms of each of the amino acids. This table can be used to refer the atom types against the atom numbers. The charge fluctuation for these atom numbers have been plotted for all the residues of the both the proteins in Supplementary Figs. 1 and 2. Population Distribution plot for umbrella sampling simulations are provided as Supplementary Fig. 3. Detailed PCA on individual trajectories have been provided. The advantages and disadvantages of the three different treatment employed in the parallel cascade MD simulations have been discussed in Supplementary Table 2 (DOCX 3297 kb)
Rights and permissions
About this article
Cite this article
Jani, V., Sonavane, U. & Joshi, R. Traversing the folding pathway of proteins using temperature-aided cascade molecular dynamics with conformation-dependent charges. Eur Biophys J 45, 463–482 (2016). https://doi.org/10.1007/s00249-016-1115-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00249-016-1115-4