Abstract
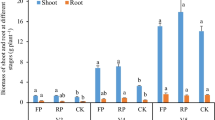
The introduction of legumes into rotations can improve nitrogen use efficiency and crop yield; however, its microbial mechanism involved remains unclear. This study aimed to explore the temporal impact of peanut introduction on microorganisms related to nitrogen metabolism in rotation systems. In this study, the dynamics of diazotrophic communities in two crop seasons and wheat yields of two rotation systems: winter wheat – summer maize (WM) and spring peanut → winter wheat – summer maize (PWM) in the North China Plain were investigated. Our results showed that peanut introduction increased wheat yield and biomass by 11.6% (p < 0.05) and 8.9%, respectively. Lower Chao1 and Shannon indexes of the diazotrophic communities were detected in soils that sampling in June compared with those sampling in September, although no difference was found between WM and PWM. Principal co-ordinates analysis (PCoA) showed that rotation system significantly changed the diazotrophic community structures (PERMANOVA; p < 0.05). Compared with WM, the genera of Azotobacter, Skermanella, Azohydromonas, Rhodomicrobium, Azospirillum, Unclassified_f_Opitutaceae, and Unclassified_f_Rhodospirillaceae were significantly enriched (p < 0.05) in PWM. Furthermore, rotation system and sampling time significantly influenced soil properties, which significantly correlated with the top 15 genera in relative abundance. Partial least squares path modeling (PLS-PM) analysis further showed that the diazotrophic community diversity (alpha- and beta-diversity) and soil properties (pH, SOC and TN) significantly affected wheat yield. In conclusion, legume inclusion has the potential to stabilize diazotrophic community structure at the temporal scales and increase subsequent crop yield.








Similar content being viewed by others
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Liu Y, Wang E, Yang X, Wang J (2010) Contributions of climatic and crop varietal changes to crop production in the North China Plain, since 1980s. Global Change Biol 16(8):2287–2299. https://doi.org/10.1111/j.1365-2486.2009.02077.x
Yu XJ, Chen Q, Shi WC, Gao Z, Sun X, Dong JJ, Li J, Wang HT, Gao JG, Liu ZG, Zhang M (2021) Interactions between phosphorus availability and microbes in a wheat-maize double cropping system: a reduced fertilization scheme. J Integr Agr 21(3):840–854. https://doi.org/10.1016/S2095-3119(20)63599-7
Ghosh PK, Hazra KK, Venkatesh MS, Nath CP, Singh J, Nadarajan N (2019) Increasing soil organic carbon through crop diversification in cereal-cereal rotations of Indo-Gangetic plain. P Natl A Sci India B 89:429–440. https://doi.org/10.1007/s40011-017-0953-x
Grażyna S, Agnieszka F, Katarzyna P, Jerzy S, Karolina R, Hanna S (2019) The long-term effect of legumes as forecrops on the productivity of rotation winter triticale-winter rape with nitrogen fertilisation. Acta Agr Scand B-S P 70(2):128–134. https://doi.org/10.1080/09064710.2019.1677766
Zhao J, Chen J, Beillouin D, Lambers H, Yang YD, Smith P, Zeng ZH, Olesen JE, Zang HD (2022) Global systematic review with meta-analysis reveals yield advantage of legume-based rotations and its drivers. Nat Commun 13:4926. https://doi.org/10.1038/s41467-022-32464-0
Congreves KA, Hayes A, Verhallen EA, Van ELL (2015) Long-term impact of tillage and crop rotation on soil health at four temperate agroecosystems. Soil Till Res 152:17–28. https://doi.org/10.1016/j.still.2015.03.012
Lasisi A, Liu K (2023) A global meta-analysis of pulse crop effect on yield, resource use, and soil organic carbon in cereal- and oilseed-based cropping systems. Field Crop Res 294:108857. https://doi.org/10.1016/j.fcr.2023.108857
Ladha JK, Peoples MB, Reddy PM, Biswas JC, Bennett A, Jat ML, Krupnik TJ (2022) Biological nitrogen fixation and prospects for ecological intensification in cereal-based cropping systems. Field Crop Res 283:108541. https://doi.org/10.1016/j.fcr.2022.108541
Pommier T, Cantarel AAM, Grigulis K, Lavorel S, Legay N, Baxendale C, Bardgett RD, Bahn M, Poly F, Cl´ement JC, (2018) The added value of including key microbial traits to determine nitrogen-related ecosystem services in managed grasslands. J Appl Ecol 55(1):49–58. https://doi.org/10.1111/1365-2664.13010
Davies-Barnard T, Friedlingstein P (2020) The global distribution of biological nitrogen fixation in terrestrial natural ecosystems. Global Biogeochem Cy 34(3):e2019GB006387. https://doi.org/10.1029/2019GB006387
Stein LY, Klotz MG (2016) The nitrogen cycle. Curr Biol 26(3):R94–R98. https://doi.org/10.1016/j.cub.2015.12.021
Kuypers MMM, Marchant HK, Kartal B (2018) The microbial nitrogen-cycling network. Nat Rev Microbiol 16(5):263–276. https://doi.org/10.1038/nrmicro.2018.9
Galloway JN, Dentener FJ, Capone DG, Boyer EW, Howarth RW, Seitzinger SP, Asner GP, Cleveland CC, Green PA, Holland EA, Karl DM, Michaels AF, Porter JH, Townsend AR, Smarty CJVR (2004) Nitrogen cycles past, present, and future. Biogeochemistry 70(2):153–226. https://doi.org/10.1007/s10533-004-0370-0
Reed SC, Cleveland CC, Townsend AR (2011) Functional ecology of free-living nitrogen fixation: a contemporary perspective. Annu Rev Ecol Evol S 42(1):489–512. https://doi.org/10.1146/annurev-ecolsys-102710-145034
Lindsay EA, Colloff MJ, Gibb NL, Wakelin SA (2010) The abundance of microbial functional genes in grassy woodlands is influenced more by soil nutrient enrichment than by recent weed invasion or livestock exclusion. Appl Environ microb 76(16):5547–5555. https://doi.org/10.1128/AEM.03054-09
Collavino MM, Tripp HJ, Frank IE, Vidoz ML, Calderoli PA, Donato M, Zehr JP, Aguilar OM (2014) nifH pyrosequencing reveals the potential for locationspecific soil chemistry to influence N2-fixing community dynamics. Environ Microbiol 16(10):3211–3223. https://doi.org/10.1111/1462-2920.12423
Berthrong ST, Yeager CM, Gallegos-Graves L, Steven B, Eichorst SA, Jackson RB, Kuske CR (2014) Nitrogen fertilization has a stronger effect on soil nitrogen-fixing bacterial communities than elevated atmospheric CO2. Appl Environ Microbiol 80(10):3103–3112. https://doi.org/10.1128/AEM.04034-13
Mergel A, Kloos K, Bothe H (2001) Seasonal fluctuations in the population of denitrifying and N2-fixing bacteria in an acid soil of a Norway spruce forest. Plant Soil 203:145–160. https://doi.org/10.1023/A:1004826116981
Hayden HL, Drake J, Imhof M, Oxley APA, Norng S, Mele PM (2010) The abundance of nitrogen cycle genes amoA and nifH depends on land-uses and soil types in South-Eastern Australia. Soil Biol Biochem 42:1774-e1783. https://doi.org/10.1016/j.soilbio.2010.06.015
Nelson DR, Mele PM (2006) The impact of crop residue amendments and lime on microbial community structure and nitrogen-fixing bacteria in the wheat rhizosphere. Aust J Soil Res 44(4):319–329. https://doi.org/10.1071/SR06022
Fan KK, Weisenhorn P, Gilbert JA, Shi Y, Bai Y, Chu HY (2018) Soil pH correlates with theco-occurrence and assemblage process of diazotrophic communities in rhizosphere and bulk soils of wheat fields. Soil Biol Biochem 121:185–192. https://doi.org/10.1016/j.soilbio.2018.03.017
Wakelin SA, Gupta VVSR, Forrester ST (2010) Regional and local factors affecting diversity, abundance and activity of free-living, N2-fixing bacteria in Australian agricultural soils. Pedobiologia 53(10):391–399. https://doi.org/10.1016/j.pedobi.2010.08.001
Xiao CH, Tang H, Pu LJ, Sun DM, Ma JZ, Yu M, Duan RS (2010) Diversity of nitrogenase (nifH) genes pool in soybean field soil after continuous and rotational cropping. J Basic Microb 50:373–379. https://doi.org/10.1002/jobm.200900317
Zou J, Yao Q, Liu J, Li YS, Song FQ, Liu XB, Wang GH (2020) Changes of diazotrophic communities in response to cropping systems in a Mollisol of Northeast China. Peer J 8:e9550. https://doi.org/10.7717/peerj.9550
Zhang X, Zhang R, Zhang C, Gao J, Wang X, Fan F, Ma X, Yin H, Feng K, Deng Y (2017) Thirty-one years of rice-rice-green manure rotations shape the rhizosphere microbial community and enrich beneficial bacteria. Soil Biol Biochem 104:208–217. https://doi.org/10.1016/j.soilbio.2016.10.023
Shen H, Yan W, Yang X, He XH, Wang X, Zhang YT, Wang B, Xia QY (2020) Co-occurrence network analyses of rhizosphere soil microbial PLFAs and metabolites over continuous cropping seasons in tobacco. Plant Soil 452(4):119–135. https://doi.org/10.1007/s11104-020-04560-x
Wu X, Jousset A, Guo S, Karlsson I, Zhao QY, Wu HS, Kowalchuk GA, Shen QR, Li R, Geisen S (2018) Soil protist communities form a dynamic hub in the soil microbiome. ISME J 12:634–638. https://doi.org/10.1038/ismej.2017.171
Zhang LB, Chen MM, Chen XW, Wang JN, Zhang Y, Xiao XL, Liu HJ, Zhang R, Xu DP, Jiao NZ, Zhang Y (2021) Nitrifiers drive successions of particulate organic matter and microbial community composition in a starved macrocosm. Environ Int 157:106776. https://doi.org/10.1016/j.envint.2021.106776
Guseva K, Darcy S, Simon E, Alteio LV, Montesinos-Navarro A, Kaiser C (2022) From diversity to complexity: Microbial networks in soils. Soil Biol Biochem 169:108604. https://doi.org/10.1016/j.soilbio.2022.108604
Wu H, Hu B, Han H, Cheng X, Kang F (2022) Network analysis reveals the regulatory effect of mixed stands on ecosystem structure and functions in the Loess Plateau. China. Sci Total Environ 824:153588. https://doi.org/10.1016/j.scitotenv.2022.153588
Hartman K, van der Heijden MG, Wittwer RA, Banerjee S, Walser JC, Schlaeppi K (2018) Cropping practices manipulate abundance patterns of root and soil microbiome members paving the way to smart farming. Microbiome 6:14. https://doi.org/10.1186/s40168-017-0389-9
Yang XL, Chen YQ, Pacenka S, Gao WS, Ma L, Wang GY, Yan P, Sui P, Steenhuis TS (2015) Effect of diversified crop rotations on groundwater levels and crop water productivity in the North China Plain. J Hydrol 522:428–438. https://doi.org/10.1016/j.jhydrol.2015.01.010
Bao S (2000) Agrochemical analysis of soil. China Agricultural Press, Beijing, China
Rosch C, Mergel A, Bothe H (2002) Biodiversity of denitrifying and dinitrogen-fixing bacteria in an acid forest soil. Appl Environ Microbiol 68(8):3818–3829. https://doi.org/10.1128/AEM.68.8.3818-3829.2002
Yang YD, Feng XM, Hu YG, Zeng ZH (2019) The diazotrophic community in oat rhizosphere: effects of legume intercropping and crop growth stage. Front Agricul Sci Engin 6(2):162–171. https://doi.org/10.15302/J-FASE-2018212
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Noah F, Antonio GP, Julia KG, Jeffrey IG, Gavin AH, Scott TK, Dan K, Jeremy EK, Ruth EL, Catherine AL, Daniel MD, Brian DM, Meg P, Jens R, Joel RS, Peter JT, William AW, Jeremy W, Tanya Y, Jesse Z, Rob K (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. https://doi.org/10.1038/nmeth.f.303
Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200. https://doi.org/10.1093/bioinformatics/btr381
Edgar RC (2013) UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10(10):996–998. https://doi.org/10.1038/NMETH.2604
Cole JR, Wang Q, Fish JA, Chai B, Mcgarrell DM, Sun Y, Brown CT, PorrasAlfaro A, Kuske CR, Tiedje JM (2014) Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucleic Acids Res 42(D1):633–642. https://doi.org/10.1093/nar/gkt1244
Fish JA, Chai B, Wang Q, Sun Y, Brown CT, Tiedje JM, Cole JR (2013) FunGene: the functional gene pipeline and repository. Front Microbiol 4(4):291. https://doi.org/10.3389/fmicb.2013.00291
Schloss PD, Gevers D, Westcott SL (2011) Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLoS One 6:e27310. https://doi.org/10.1371/JOURNAL.PONE.0027310
R Core Team (2014) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna (Austria)
Yu TB, Cheng L, Liu Q, Wang SS, Zhou Y, Zhong HB, Tang MF, Nian H, Lian TX (2022) Effects of waterlogging on soybean rhizosphere bacterial community using V4, LoopSeq, and PacBio 16S rRNA sequence. Microbiol Spectr 10(1):e02011-e2021. https://doi.org/10.1128/spectrum.0201
Yu JJ, Li S, Li R, Zhang J, Liu YH, Lv LF, Zhu H, Wu WL, Li WL (2017) Plant cultivars imprint the rhizosphere bacterial community composition and association networks. Soil Biol Biochem 109:145–155. https://doi.org/10.1016/j.soilbio.2017.02.010
Delgado-Baquerizo M, Reith F, Dennis PG, Hamonts K, Powell JR, Young A, Singh BK, Bissett A (2018) Ecological drivers of soil microbial diversity and soil biological networks in the Southern Hemisphere. Ecology 99:583–596. https://doi.org/10.1002/ecy.2137
Tenenhaus M, Vinzi VE, Chatelin YM, Lauro C (2005) PLS path modeling. Comput Stat Data Anal 48:159–205. https://doi.org/10.1016/j.csda.2004.03.005
Gao X, Chen H, Govaert L, Wang WP, Yang J (2019) Responses of zooplankton body size and community trophic structure to temperature change in a subtropical reservoir. Ecol Evol 9(22):12544–12555. https://doi.org/10.1002/ece3.5718
Zhang YP, Li W, Lu P, Xu TY, Pan K (2022) Three preceding crops increased the yield of and inhibited clubroot disease in continuously monocropped chinese cabbage by regulating the soil properties and rhizosphere microbial community. Microorganisms 10:799. https://doi.org/10.3390/microorganisms10040799
Brophy LS, Heichel GH (1989) Nitrogen release from roots of alfalfa and soybean grown in sand culture. Plant Soil 116(1):77–84. https://doi.org/10.1007/BF02327259
Plaza-Bonilla D, Nolot JM, Raffaillac D, Justes E (2017) Innovative cropping systems to reduce N inputs and maintain wheat yields by inserting grain legumes and cover crops in southwestern France. Eur J Agron 82:331–341. https://doi.org/10.1016/j.eja.2016.05.010
Liu N, Li Y, Cong P, Wang J, Guo W, Pang H, Zhang L (2021) Depth of straw incorporation significantly alters crop yield, soil organic carbon and total nitrogen in the North China Plain. Soil Till Res 205:104772. https://doi.org/10.1016/j.still.2020.104772
Guinet M, Nicolardot B, Voisin AS (2020) Nitrogen benefits of ten legume pre-crops for wheat assessed by field measurements and modelling. Eur J Agron 120:126–151. https://doi.org/10.1016/j.eja.2020.126151
Prusiński J, Borowska M, Kaszkowiak E, Olszak G (2016) The after-effect of chosen Fabaceae forecrops on the yield of grain and protein in winter triticale (Triticosecale sp. Wittmack ex A. Camus 1927) fertilized with mineral nitrogen. Plant Soil Environ 62:571–576. https://doi.org/10.17221/463/2016-PSE
Zhao J, Yanga YD, Zhang K, Jeong J, Zeng ZH, Zang HD (2020) Does crop rotation yield more in China? A meta-analysis. Field Crop Res 245:107659. https://doi.org/10.1016/j.fcr.2019.107659
Faligowska A, Szymańska G, Panasiewicz K, Szukała J, Koziara W, Ratajczak K (2019) The long-term effect of legumes as forecrops on the productivity of rotation (winter rape-winter wheat-winter wheat) with nitrogen fertilization. Plant Soil Enviro 65:138–144. https://doi.org/10.17221/556/2018-PSE
Zang H, Qian X, Wen Y, Hu Y, Ren C, Zeng Z, Guo L, Wang C (2018) Contrasting carbon and nitrogen rhizodeposition patterns of soya bean (Glycine max L.) and oat (Avena nuda L.). Eur J Soil Sci 69(4):625–633. https://doi.org/10.1111/ejss.12811
Meng PP, Liu X, Qiu HZ, Zhang WR, Zhang CH, Wang D, Zhang JL, Shen QR (2012) Fungal population structure and its biological effect in rhizosphere soil of continuously cropped potato. Chinese J Appl Ecol 23(11):3079–3086
Li ZL, Zeng ZQ, Tian DS, Wang JS, Wang BX, Chen HYH, Quan Q, Chen WN, Yang JL, Meng C, Wang Y, Niu S (2020) Global variations and controlling factors of soil nitrogen turnover rate. Earth-Sci Rev 207:103250. https://doi.org/10.1016/j.earscirev.2020.103250
Gabriel YK, Moinet RH, van Vuuren DP, Giller KE (2023) Carbon for soils, not soils for carbon. Glob Change Biol 29:2384–2398. https://doi.org/10.1111/gcb.16570
Liu J, Yu Z, Yao Q, Hu X, Zhang W, Mi G, Chen X, Wang G (2017) Distinct soil bacterial communities in response to the cropping system in a Mollisol of northeast China. Appl Soil Ecol 119:407–416. https://doi.org/10.1016/j.apsoil.2017.07.013
Yin C, Jones KL, Peterson DE, Garrett KA, Hulbert SH, Paulitz TC (2010) Members of soil bacterial communities sensitive to tillage and crop rotation. Soil Biol Biochem 42(12):2111–2118. https://doi.org/10.1016/j.soilbio.2010.08.006
Fierer N, Jackson RB (2006) The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci USA 103:626–631. https://doi.org/10.1073/pnas.0507535103
Zhou D, Huang X (2015) Root and bacterial secretions regulate the interaction between plants and PGPR leading to distinct plant growth promotion effects. Plant Soil 401:259–272. https://doi.org/10.1007/s11104-015-2743-7
Sheridan LW, Francesca DF, Maurizio Z, Albert V, Pierre H, Giuliano B (2022) Pea-wheat rotation affects soil microbiota diversity, community structure, and soil borne pathogens. Microorganisms 10:370. https://doi.org/10.3390/microorganisms10020370
Ridl J, Kolar M, Strejcek M, Strand H, Stursa P, Paces J, Macek T, Uhlik O (2016) Plants rather than mineral fertilization shape microbial community structure and functional potential in legacy contaminated soil. Front Microbiol 7(837):995. https://doi.org/10.3389/fmicb.2016.00995
Paungfoo-Lonhienne C, Wang W, Yeoh YK, Halpin N (2017) Legume crop rotation suppressed nitrifying microbial community in a sugarcane cropping soil. Sci Rep 7(1):16707. https://doi.org/10.1038/s41598-017-17080-z
Garrido-Oter R, Nakano RT, Dombrowski N, Ma KW, AgBiome T, McHardy AC, Schulze-Lefert P (2018) Modular traits of the rhizobiales root microbiota and their evolutionary relationship with symbiotic rhizobia. Cell Host Microbe 24:155–167. https://doi.org/10.1016/j.chom.2018.06.006
Han Q, Ma Q, Chen Y, Tian B, Xu LX, Bai Y, Chen WF, Li X (2020) Variation in rhizosphere microbial communities and its association with the symbiotic efficiency of rhizobia in soybean. ISME J 14:1915–1928. https://doi.org/10.1038/s41396-020-0648-9
Sul WJ, Asuming-Brempong S, Wang Q, Tourlousse DM, Penton CR, Deng Y, Rodrigues JLM, Adiku SGK, Jones JW, Zhou J, Cole JR, Tiedje JM (2013) Tropical agricultural land management influences on soil microbial communities through its effect on soil organic carbon. Soil Biol Biochem 65:33–38. https://doi.org/10.1016/j.soilbio.2013.05.007
Bürgmann H, Meier S, Bunge M, Widmer F, Zeyer J (2005) Effects of model root exudates on structure and activity of a soil diazotroph community. Environ Microbiol 7(11):1711–1724. https://doi.org/10.1111/j.1462-2920.2005.00818.x
Bulgarelli D, Schlaeppi K, Spaepen S, Loren V, van Temaat E, Schulze-Lefert P (2013) Structure and functions of the bacterial microbiota of plants. Annu Rev Plant Biol 64:807–838. https://doi.org/10.1146/annurev-arplant-050312-1201
Song Y, Xu M, Li XN, Bian YR, Wang F, Yang XL, Gu CG, Jiang X (2018) Long-term plastic greenhouse cultivation changes soil microbial community structures: a case study. J Agr Food Chem 66(34):8941–8948. https://doi.org/10.1021/acs.jafc.8b01829
Sandipan S, Radomir S, Nicole ET, Kate S (2021) Adding alfalfa to an annual crop rotation shifts the composition and functional responses of tomato rhizosphere microbial communities. Appl Soil Ecol 167:104102. https://doi.org/10.1016/j.apsoil.2021.104102
Fazal A, Yang M, Wen Z, Ali F, Ren R, Hao C, Chen XY, Fu JY, Wang X, Jie WC, Yin TM, Lu GH, Qi JL, Yang YH (2021) Differential microbial assemblages associated with shikonin-producing borage species in two distinct soil types. Sci Rep 11:10788. https://doi.org/10.1038/s41598-021-9
Sun A, Jiao XY, Chen Q, Trivedi P, Li Z, Li F, Zheng Y, Lin YX, Hu HW, He JZ (2021) Fertilization alters protistan consumers and parasites in crop-associated microbiomes. Environ Microbiol 23:2169–2183. https://doi.org/10.1111/1462-2920.15385
Chen SM, Waghmode TR, Sun RB, Kuramae EE, Hu CS, Liu BB (2019) Root-associated microbiomes of wheat under the combined effect of plant development and nitrogen fertilization. Microbiome 7:136. https://doi.org/10.1186/s40168-019-0750-2
Sumbul A, Ansari RA, Rizvi R, Mahmood I (2020) Azotobacter: A potential bio-fertilizer for soil and plant health management. Saudi J Biol Sci 27:3634–3640. https://doi.org/10.1016/j.sjbs.2020.08.004
Lladó S, Jiménez N, Viñas M, Solanas AM (2009) Microbial populations related to PAH biodegradation in an aged biostimulated creosote-contaminated soil. Biodegradation 20:593–601. https://doi.org/10.1007/s10532-009-9247-1
Zhu W, Huang J, Li M, Li X, Wang G (2014) Genomic analysis of Skermanella stibiiresistens type strain SB22T. Stand Geonomic Scie 9(3):1211–1220. https://doi.org/10.4056/sigs.5751047
Faust K, Raes J (2012) Microbial interactions: from networks to models. Nat Rev Microbiol 10:538–550. https://doi.org/10.1038/nrmicro2832
Song Y, Wilson AJ, Zhang XC, Thoms D, Sohrab R, Song SY, Geissmann Q, Liu Y, Walgren L, He SY, Haney CH (2021) FERONIA restricts Pseudomonas in the rhizosphere microbiome via regulation of reactive oxygen species. Nat Plants 7:644–654. https://doi.org/10.1038/s41477-021-00914-0
Coyte KZ, Schluter J, Foster KR (2015) The ecology of the microbiome: networks, competition, and stability. Science 350:663–666. https://doi.org/10.1126/science.aad2602
Kudjordjie EN, Sapkota R, Steffensen SK, Fomsgaard IS, Nicolaisen M (2019) Maize synthesized benzoxazinoids affect the host associated microbiome. Microbiome 7:59. https://doi.org/10.1186/s40168-019-0677-7
Mills MM, Turk-Kubo KA, van Dijken GL, Henke BA, Harding K, Wilson ST, Kevin RA, Zehr JP (2020) Unusual marine cyanobacteria/haptophyte symbiosis relies on N2 fixation even in N-rich environments. ISME J 14(10):2395–2406. https://doi.org/10.1038/s41396-020-0691-6
Wang C, Wang B, Zheng M, Song W, Wen S, Zhu C, Shen R (2017) Impact of 25 years of inorganic fertilization on diazotrophic abundance and community structure in an acidic soil in southern China. Soil Biol Biochem 113:240–249. https://doi.org/10.1016/j.soilbio.2017.06.019
Xu Y, Wang T, Li H, Ren C, Chen J, Yang G, Han XH, Feng YZ, Ren GG, Wang X (2019) Variations of soil nitrogen-fixing microorganism communities and nitrogen fractions in a Robinia pseudoacacia chronosequence on the Loess Plateau of China. CATENA 174:316–323. https://doi.org/10.1016/j.catena.2018.11.009
Poly F, Ranjard L, Nazaret S, Gourbière F, Monrozier LJ (2001) Comparison of nifH gene pools in soils and soil microenvironments with contrasting properties. Appl Environ Microbiol 67:2255–2262. https://doi.org/10.1128/AEM.67.5.2255-2262.2001
Gamalero E, Glick BR (2019) Plant growth-promoting bacteria in agricultural and stressed soils. In: van Elsas JD, Trevors JT, Rosado AS, Nannipieri P (eds) Modern soil microbiology, 3rd edn. CRC Press, Boca Raton, pp 361–380
Wurzburger N, Bellenger JP, Kraepiel AML, Hedin LO (2012) Molybdenum and phosphorus interact to constrain asymbiotic nitrogen fixation in tropical forests. PLoS One 7:e33710. https://doi.org/10.1371/journal.pone.0033710
Huang JF, Pang YW, Zhang FB, Huang QY, Zhang M, Tang SH, Fu HT, Li P (2019) Suppression of Fusarium wilt of banana by combining acid soil ameliorant with biofertilizer made from Bacillus velezensis H-6. Eur J Plant Pathol 154:585–596. https://doi.org/10.1007/s10658-019-01683-5
Liu ZX, Liu JJ, Yu ZH, Lia YS, Hua XJ, Gua HD, Lia LJ, Jina J, Liua XB, Wang GH (2022) Archaeal communities perform an important role in maintaining microbial stability under long term continuous cropping systems. Sci Total Environ 838(3):156413. https://doi.org/10.1016/j.scitotenv.2022.156413
Oldroyd GED, Leyser O (2020) A plant’s diet, surviving in a variable nutrient environment. Science 368:6486. https://doi.org/10.1126/science.aba0196
Fan KK, Delgado-Baquerizo M, Guo X, Wang DZ, Zhu YG, Chu HY (2021) Biodiversity of key-stone phylotypes determines crop production in a 4-decade fertilization experiment. ISME J 15:550–561. https://doi.org/10.1038/s41396-020-00796-8
Garland G, Edlinger A, Banerjee S, Degrune F, García-Palacios P, Pescador DS, Herzog C, Romdhane S, Saghai A, Spor A, Wagg C, Hallin S, Maestre FT, Philippot L, Rillig MC, Marcel GA (2021) Crop cover is more important than rotational diversity for soil multifunctionality and cereal yields in European cropping systems. Nat Food 2:28–37. https://doi.org/10.1038/s43016-020-00210-8
Weidner S, Koller R, Latz E, Kowalchuk G, Bonkowski M, Scheu S, Jousset A (2015) Bacterial diversity amplifies nutrientbased plant-soil feedbacks. Funct Ecol 29:1341–1349. https://doi.org/10.1111/1365-2435.12445
Suzuki C, Kunito T, Aono T, Liu CT, Oyaizu H (2005) Microbial indices of soil fertility. J Appl Microbiol 98:1062–1074. https://doi.org/10.1111/j.1365-2672.2004.02529.x
Funding
This work was financial supported by the National Science Foundation of China (31901470) and the National Key Research and Development Program of China (2016YFD0300205-01).
Author information
Authors and Affiliations
Contributions
YYD and ZZH conceived and designed the research, acquired the funding. YTB and NJW conducted experiments. YTB and NJW analyzed the data. YTB and NJW wrote the paper. YYD, ZHD and ZZH revised the paper. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Ethics approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Conflict of interest
All authors declare no conflict of this manuscript.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yu, T., Nie, J., Zang, H. et al. Peanut-based Rotation Stabilized Diazotrophic Communities and Increased Subsequent Wheat Yield. Microb Ecol 86, 2447–2460 (2023). https://doi.org/10.1007/s00248-023-02254-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-023-02254-2