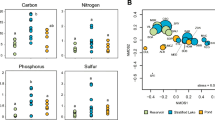
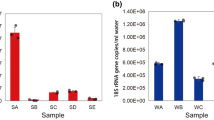
Abstract
Microeukaryotic diversity, community structure, and their regulating mechanisms remain largely unclear in chemosynthetic ecosystems. Here, using high-throughput sequencing data of 18S rRNA genes, we explored microeukaryotic communities from the Haima cold seep in the northern South China Sea. We compared three distinct habitats: active, less active, and non-seep regions, with vertical layers (0–25 cm) from sediment cores. The results showed that seep regions harbored more abundant and diverse parasitic microeukaryotes (e.g., Apicomplexa and Syndiniales) as indicator species, compared to nearby non-seep region. Microeukaryotic community heterogeneity was larger between habitats than within habitat, and greatly increased when considering molecular phylogeny, suggesting the local diversification in cold-seep sediments. Microeukaryotic α-diversity at cold seeps was positively increased by metazoan richness and dispersal rate of microeukaryotes, while its β-diversity was promoted by heterogeneous selection mainly from metazoan communities (as potential hosts). Their combined effects led to the significant higher γ-diversity (i.e., total diversity in a region) at cold seeps than non-seep regions, suggesting cold-seep sediment as a hotspot for microeukaryotic diversity. Our study highlights the importance of microeukaryotic parasitism in cold-seep sediment and has implications for the roles of cold seep in maintaining and promoting marine biodiversity.








Similar content being viewed by others
Data Availability
Raw sequencing reads for analyses in this study were deposited in online open database of National Center for Biotechnology Information Search database (NCBI) with projection accession number of PRJNA849592 (submission ID: SUB11547907, to be released in June 2023). R scripts used for analysis can be found from the website: https://github.com/xzhimenghkust/Haima-cold-seep-microeukaryotes-Rscripts.
References
Boetius A, Wenzhöfer F (2013) Seafloor oxygen consumption fuelled by methane from cold seeps. Nat Geosci 6:725–734. https://doi.org/10.1038/ngeo1926
Valentine DL, Blanton DC, Reeburgh WS, Kastner M (2001) Water column methane oxidation adjacent to an area of active hydrate dissociation, Eel River Basin. Geochim Cosmochim Acta 65:2633–2640. https://doi.org/10.1016/S0016-7037(01)00625-1
Sommer S, Pfannkuche O, Linke P et al (2006) Efficiency of the benthic filter: biological control of the emission of dissolved methane from sediments containing shallow gas hydrates at Hydrate Ridge. Global Biogeochem Cycles 20. https://doi.org/10.1029/2004GB002389
Li Z, Pan D, Wei G et al (2021) Deep sea sediments associated with cold seeps are a subsurface reservoir of viral diversity. ISME J 15:2366–2378. https://doi.org/10.1038/s41396-021-00932-y
Ruff SE, Biddle JF, Tesked AP et al (2015) Global dispersion and local diversification of the methane seep microbiome. Proc Natl Acad Sci U S A 112:4015–4020. https://doi.org/10.1073/pnas.1421865112
van Dover CL, German CR, Speer KG et al (2002) Evolution and biogeography of deep-sea vent and seep invertebrates. Science (1979) 295:1253–1257 https://doi.org/10.1126/science.1067361
Ristova PP, Wenzhöfer F, Ramette A et al (2015) Spatial scales of bacterial community diversity at cold seeps (Eastern Mediterranean Sea). ISME J 9:1306–1318. https://doi.org/10.1038/ismej.2014.217
Vanreusel A, Andersen AC, Boetius A et al (2009) Biodiversity of cold seep ecosystems along the European margins. Oceanography 22:110–127. https://doi.org/10.2307/24860929
Xu T, Wang Y, Sun J et al (2021) Hidden historical habitat-linked population divergence and contemporary gene flow of a deep-sea patellogastropod limpet. Mol Biol Evol 38:5640–5654. https://doi.org/10.1093/molbev/msab278
Bar-On YM, Phillips R, Milo R (2018) The biomass distribution on Earth. Proc Natl Acad Sci U S A 115:6506–6511. https://doi.org/10.1073/pnas.1711842115
Burki F, Sandin MM, Jamy M (2021) Diversity and ecology of protists revealed by metabarcoding. Curr Biol 31:R1267–R1280. https://doi.org/10.1016/j.cub.2021.07.066
Rodríguez-Martínez R, Leonard G, Milner DS et al (2020) Controlled sampling of ribosomally active protistan diversity in sediment-surface layers identifies putative players in the marine carbon sink. ISME J 14:984–998. https://doi.org/10.1038/s41396-019-0581-y
de Vargas C, Audic S, Henry N et al (1979) (2015) Eukaryotic plankton diversity in the sunlit ocean. Science 348:1261605. https://doi.org/10.1126/science.1261605
Hu SK, Campbell V, Connell P et al (2016) Protistan diversity and activity inferred from RNA and DNA at a coastal ocean site in the eastern North Pacific. FEMS Microbiol Ecol 92. https://doi.org/10.1093/femsec/fiw050
Zhu F, Massana R, Not F et al (2005) Mapping of picoeucaryotes in marine ecosystems with quantitative PCR of the 18S rRNA gene. FEMS Microbiol Ecol 52:79–92. https://doi.org/10.1016/j.femsec.2004.10.006
Gong J, Dong J, Liu X, Massana R (2013) Extremely high copy numbers and polymorphisms of the rDNA operon estimated from single cell analysis of oligotrich and peritrich ciliates. Protist 164:369–379. https://doi.org/10.1016/j.protis.2012.11.006
Takishita K, Kakizoe N, Yoshida T, Maruyama T (2010) Molecular evidence that phylogenetically diverged ciliates are active in microbial mats of deep-sea cold-seep sediment. J Eukaryot Microbiol 57:76–86. https://doi.org/10.1111/j.1550-7408.2009.00457.x
Takishita K, Tsuchiya M, Reimer JD, Maruyama T (2006) Molecular evidence demonstrating the basidiomycetous fungus Cryptococcus curvatus is the dominant microbial eukaryote in sediment at the Kuroshima Knoll methane seep. Extremophiles 10:165–169. https://doi.org/10.1007/s00792-005-0495-7
Takishita K, Yubuki N, Kakizoe N et al (2007) Diversity of microbial eukaryotes in sediment at a deep-sea methane cold seep: surveys of ribosomal DNA libraries from raw sediment samples and two enrichment cultures. Extremophiles 11:563–576. https://doi.org/10.1007/s00792-007-0068-z
Nagahama T, Takahashi E, Nagano Y et al (2011) Molecular evidence that deep-branching fungi are major fungal components in deep-sea methane cold-seep sediments. Environ Microbiol 13:2359–2370. https://doi.org/10.1111/j.1462-2920.2011.02507.x
Pasulka AL, Levin LA, Steele JA et al (2016) Microbial eukaryotic distributions and diversity patterns in a deep-sea methane seep ecosystem. Environ Microbiol 18:3022–3043. https://doi.org/10.1111/1462-2920.13185
Logares R, Deutschmann IM, Junger PC et al (2020) Disentangling the mechanisms shaping the surface ocean microbiota. Microbiome 8:1–17. https://doi.org/10.1186/s40168-020-00827-8
Wu W, Lu HP, Sastri A et al (2018) Contrasting the relative importance of species sorting and dispersal limitation in shaping marine bacterial versus protist communities. ISME J 12:485–494. https://doi.org/10.1038/ismej.2017.183
Xu Z, Cheung S, Endo H et al (2022) Disentangling the ecological processes shaping the latitudinal pattern of phytoplankton communities in the Pacific Ocean. mSystems 7:e01203–21. https://doi.org/10.1128/msystems.01203-21
O’Hara SCM, Dando PR, Schuster U et al (1995) Gas seep induced interstitial water circulation: observations and environmental implications. Cont Shelf Res 15:931–948. https://doi.org/10.1016/0278-4343(95)80003-V
Feng D, Qiu J-W, Hu Y et al (2018) Cold seep systems in the South China Sea: an overview. J Asian Earth Sci 168:3–16. https://doi.org/10.1016/j.jseaes.2018.09.021
Dini-Andreote F, Stegen JC, van Elsas JD, Salles JF (2015) Disentangling mechanisms that mediate the balance between stochastic and deterministic processes in microbial succession. Proc Natl Acad Sci U S A 112:E1326–E1332. https://doi.org/10.1073/pnas.1414261112
Nemergut DR, Schmidt SK, Fukami T et al (2013) Patterns and processes of microbial community assembly. Microbiol Mol Biol Rev 77:342–356. https://doi.org/10.1128/MMBR.00051-12
Stegen JC, Lin X, Konopka AE, Fredrickson JK (2012) Stochastic and deterministic assembly processes in subsurface microbial communities. ISME J 6:1653–1664. https://doi.org/10.1038/ismej.2012.22
Zhou J, Ning D (2017) Stochastic community assembly: does it matter in microbial ecology? Microbiol Mol Biol Rev 81:e00002-17. https://doi.org/10.1128/MMBR
Vellend M (2010) Conceptual synthesis in community ecology. Q R Biol 85:183–206. https://doi.org/10.1086/652373
Chase JM, Myers JA (2011) Disentangling the importance of ecological niches from stochastic processes across scales. Philos Trans Royal Soc B: Biol Sci 366:2351–2363. https://doi.org/10.1098/rstb.2011.0063
Hubbell SP (2011) The unified neutral theory of biodiversity and biogeography (MPB-32). Princeton Univ Press. https://doi.org/10.1515/9781400837526
Connor EF, Mccoy ED (1979) The statistics and biology of the species-area relationship. Am Nat 113. https://doi.org/10.1086/283438
Mouquet N, Loreau M (2003) Community patterns in source-sink metacommunities. Am Nat 162:544–557. https://doi.org/10.1086/378857
Wu W, Liu H (2018) Disentangling protist communities identified from DNA and RNA surveys in the Pearl River-South China Sea Continuum during the wet and dry seasons. Mol Ecol 27:4627–4640. https://doi.org/10.1111/mec.14867
Logares R, Tesson SVM, Canbäck B et al (2018) Contrasting prevalence of selection and drift in the community structuring of bacteria and microbial eukaryotes. Environ Microbiol 20:2231–2240. https://doi.org/10.1111/1462-2920.14265
Evans S, Martiny JBH, Allison SD (2017) Effects of dispersal and selection on stochastic assembly in microbial communities. ISME J 11:176–185. https://doi.org/10.1038/ismej.2016.96
Stegen JC, Lin X, Fredrickson JK et al (2013) Quantifying community assembly processes and identifying features that impose them. ISME J 7:2069–2079. https://doi.org/10.1038/ismej.2013.93
Liang Q, Hu Y, Feng D et al (2017) Authigenic carbonates from newly discovered active cold seeps on the northwestern slope of the South China Sea: constraints on fluid sources, formation environments, and seepage dynamics. Deep Sea Res 1 Oceanogr Res Pap 124:31–41. https://doi.org/10.1016/j.dsr.2017.04.015
Drummond AJ, Newcomb RD, Buckley TR et al (2015) Evaluating a multigene environmental DNA approach for biodiversity assessment. Gigascience 4:s13742–s14015. https://doi.org/10.1186/s13742-015-0086-1
Li S-P, Wang P, Chen Y et al (2020) Island biogeography of soil bacteria and fungi: similar patterns, but different mechanisms. ISME J 14:1886–1896. https://doi.org/10.1038/s41396-020-0657-8
Seeberg-Elverfeldt J, Schlüter M, Feseker T, Kölling M (2005) Rhizon sampling of porewaters near the sediment-water interface of aquatic systems. Limnol Oceanogr: Methods 3(8):361–371. https://doi.org/10.4319/lom.2005.3.361
Elwood HJ, Olsen GJ, Sogin ML (1985) The small-subunit ribosomal RNA gene sequences from the hypotrichous ciliates Oxytricha nova and Stylonychia pustulata. Mol Biol Evol 2:399–410. https://doi.org/10.1093/oxfordjournals.molbev.a040362
Bolyen E, Rideout JR, Dillon MR et al (2019) Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 37:852–857. https://doi.org/10.1038/s41587-019-0209-9
Quast C, Pruesse E, Yilmaz P et al (2012) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596. https://doi.org/10.1093/nar/gks1219
Team RC (2020) R: a language and environment for statistical computing (R Version 4.0. 2 R Foundation for Statistical Computing, Vienna, Austria). https://www.R-project.org/
Oksanen J, Blanchet FG, Kindt R et al (2013) Package ‘vegan.’ Community Ecol Packag, Version 2:1–295
Hamilton NE, Ferry M (2018) Ggtern: ternary diagrams using ggplot2. J Stat Softw 87:1–17. https://doi.org/10.18637/jss.v087.c03
Schneider LK, Anestis K, Mansour J et al (2020) A dataset on trophic modes of aquatic protists. Biodivers Data J 8:1–13. https://doi.org/10.3897/BDJ.8.e56648
Flynn KJ, Mitra A, Anestis K et al (2019) Mixotrophic protists and a new paradigm for marine ecology: where does plankton research go now? J Plankton Res 41:375–391. https://doi.org/10.1093/plankt/fbz026
de Caceres, Florian Jansen, de Caceres MM (2016) Package “indicspecies.” indicators 8
Chao A (1984) Nonparametric estimation of the number of classes in a population. Scand J Stat 11:265–270 https://www.jstor.org/stable/4615964
Faith DP (1992) Conservation evaluation and phylogenetic diversity. Biol Conserv 61:1–10. https://doi.org/10.1016/0006-3207(92)91201-3
Hardy OJ, Senterre B (2007) Characterizing the phylogenetic structure of communities by an additive partitioning of phylogenetic diversity. J Ecol 95:493–506. https://doi.org/10.1111/j.1365-2745.2007.01222.x
Legendre P, de Cáceres M (2013) Beta diversity as the variance of community data: dissimilarity coefficients and partitioning. Ecol Lett 16:951–963. https://doi.org/10.1111/ele.12141
Lozupone C, Lladser ME, Knights D et al (2011) UniFrac: an effective distance metric for microbial community comparison. ISME J 5:169–172. https://doi.org/10.1038/ismej.2010.133
Gotelli NJ (2001) Research frontiers in null model analysis. Glob Ecol Biogeogr 10:337–343. https://doi.org/10.1046/j.1466-822X.2001.00249.x
Wood S (2012) mgcv: mixed GAM computation vehicle with GCV/AIC/REML smoothness estimation
Sloan WT, Woodcock S, Lunn M et al (2007) Modeling taxa-abundance distributions in microbial communities using environmental sequence data. Microb Ecol 53:443–455. https://doi.org/10.1007/s00248-006-9141-x
Sloan WT, Lunn M, Woodcock S et al (2006) Quantifying the roles of immigration and chance in shaping prokaryote community structure. Environ Microbiol 8:732–740. https://doi.org/10.1111/j.1462-2920.2005.00956.x
Burns AR, Stephens WZ, Stagaman K et al (2016) Contribution of neutral processes to the assembly of gut microbial communities in the zebrafish over host development. ISME J 10:655–664. https://doi.org/10.1038/ismej.2015.142
Kembel SW, Cowan PD, Helmus MR et al (2010) Picante: R tools for integrating phylogenies and ecology. Bioinformatics 26:1463–1464. https://doi.org/10.1093/bioinformatics/btq166
Perkins AK, Rose AL, Grossart H-P et al (2021) Oxic and anoxic organic polymer degradation potential of endophytic fungi from the marine macroalga, Ecklonia radiata. Front Microbiol 12. https://doi.org/10.3389/fmicb.2021.726138
Taylor DL, Hollingsworth TN, McFarland JW et al (2014) A first comprehensive census of fungi in soil reveals both hyperdiversity and fine-scale niche partitioning. Ecol Monogr 84:3–20. https://doi.org/10.1890/12-1693.1
Zhang Y, Huang N, Wang M et al (2021) Microbial eukaryotes associated with sediments in deep-sea cold seeps. Front Microbiol 12:782004. https://doi.org/10.3389/fmicb.2021.782004
Mahé F, de Vargas C, Bass D et al (2017) Parasites dominate hyperdiverse soil protist communities in Neotropical rainforests. Nat Ecol Evol 1:1–8. https://doi.org/10.1038/s41559-017-0091
Moreira D, López-Garcı́a P (2003) Are hydrothermal vents oases for parasitic protists? Trends Parasitol 19:556–558. https://doi.org/10.1016/j.pt.2003.09.013
Sauvadet A, Gobet A, Guillou L (2010) Comparative analysis between protist communities from the deep-sea pelagic ecosystem and specific deep hydrothermal habitats. Environ Microbiol 12:2946–2964. https://doi.org/10.1111/j.1462-2920.2010.02272.x
Suter EA, Pachiadaki M, Taylor GT, Edgcomb VP (2022) Eukaryotic parasites are integral to a productive microbial food web in oxygen-depleted waters. Front Microbiol 12:4166. https://doi.org/10.3389/fmicb.2021.764605
del Campo J, Heger TJ, Rodríguez-Martínez R et al (2019) Assessing the diversity and distribution of apicomplexans in host and free-living environments using high-throughput amplicon data and a phylogenetically informed reference framework. Front Microbiol 2373. https://doi.org/10.3389/fmicb.2019.02373
Rueckert S, Simdyanov TG, Aleoshin VV, Leander BS (2011) Identification of a divergent environmental DNA sequence clade using the phylogeny of gregarine parasites (Apicomplexa) from crustacean hosts. PLoS One 6:e18163. https://doi.org/10.1371/journal.pone.0018163
Wakeman KC, Leander BS (2013) Identity of environmental DNA sequences using descriptions of four novel marine gregarine parasites, Polyplicarium n. gen. (Apicomplexa), from capitellid polychaetes. Mar Biodivers 43:133–147. https://doi.org/10.1007/s12526-012-0140-5
Faith DP, Baker AM (2006) Phylogenetic diversity (PD) and biodiversity conservation: some bioinformatics challenges. Evol Bioinforma 2:117693430600200000. https://doi.org/10.1177/117693430600200007
Auguet JC, Barberan A, Casamayor EO (2010) Global ecological patterns in uncultured Archaea. ISME J 4:182–190. https://doi.org/10.1038/ismej.2009.109
Murdock SA, Juniper SK (2019) Hydrothermal vent protistan distribution along the Mariana arc suggests vent endemics may be rare and novel. Environ Microbiol 21:3796–3815. https://doi.org/10.1111/1462-2920.14729
Dick GJ (2019) The microbiomes of deep-sea hydrothermal vents: distributed globally, shaped locally. Nat Rev Microbiol 17:271–283. https://doi.org/10.1038/s41579-019-0160-2
Wang Y, Zhang WP, Cao HL et al (2014) Diversity and distribution of eukaryotic microbes in and around a brine pool adjacent to the Thuwal cold seeps in the Red Sea. Front Microbiol 5:1–10. https://doi.org/10.3389/fmicb.2014.00037
Levin LA, Mendoza GF, Grupe BM et al (2015) Biodiversity on the rocks: macrofauna inhabiting authigenic carbonate at Costa Rica methane seeps. PLoS One 10. https://doi.org/10.1371/journal.pone.0131080
Wu W, Huang B (2019) Protist diversity and community assembly in surface sediments of the South China Sea. Microbiologyopen 8:e891. https://doi.org/10.1002/mbo3.891
Powell JR, Karunaratne S, Campbell CD et al (2015) Deterministic processes vary during community assembly for ecologically dissimilar taxa. Nat Commun 6:1–10. https://doi.org/10.1038/ncomms9444
Schrader J, Moeljono S, Keppel G, Kreft H (2019) Plants on small islands revisited: the effects of spatial scale and habitat quality on the species–area relationship. Ecography 42:1405–1414. https://doi.org/10.1111/ecog.04512
Orsi W, Edgcomb V, Jeon S et al (2011) Protistan microbial observatory in the Cariaco Basin, Caribbean. II. Habitat specialization. ISME Journal 5:1357–1373. https://doi.org/10.1038/ismej.2011.7
Edgcomb VP, Kysela DT, Teske A et al (2002) Benthic eukaryotic diversity in the Guaymas Basin hydrothermal vent environment. Proc Natl Acad Sci 99:7658–7662. https://doi.org/10.1073/pnas.062186399
Meier DV, Pjevac P, Bach W et al (2017) Niche partitioning of diverse sulfur-oxidizing bacteria at hydrothermal vents. ISME J 11:1545–1558. https://doi.org/10.1038/ismej.2017.37
Zhu P, Wang Y, Shi T et al (2018) Genetic diversity of benthic microbial eukaryotes in response to spatial heterogeneity of sediment geochemistry in a mangrove ecosystem. Estuaries Coasts 41:751–764. https://doi.org/10.1007/s12237-017-0317-z
Orsi WD, Richards TA, Francis WR (2017) Predicted microbial secretomes and their target substrates in marine sediment. Nat Microbiol 3:32–37. https://doi.org/10.1038/s41564-017-0047-9
Matthiessen B, Mielke E, Sommer U (2010) Dispersal decreases diversity in heterogeneous metacommunities by enhancing regional competition. Ecology 91:2022–2033. https://doi.org/10.1890/09-1395.1
Warren PH (1996) The effects of between-habitat dispersal rate on protist communities and metacommunities in microcosms at two spatial scales. Oecologia 105:132–140. https://doi.org/10.1007/BF00328801
Brown JH, Kodric-Brown A (1977) Turnover rates in insular biogeography: effect of immigration on extinction. Ecology 58:445–449. https://doi.org/10.2307/1935620
Zha Y, Berga M, Comte J, Langenheder S (2016) Effects of dispersal and initial diversity on the composition and functional performance of bacterial communities. PLoS One 11:e0155239. https://doi.org/10.1371/journal.pone.0155239
Shen D, Langenheder S, Jürgens K (2018) Dispersal modifies the diversity and composition of active bacterial communities in response to a salinity disturbance. Front Microbiol 9:2188. https://doi.org/10.3389/fmicb.2018.02188
Danovaro R, Snelgrove PVR, Tyler P (2014) Challenging the paradigms of deep-sea ecology. Trends Ecol Evol 29:465–475. https://doi.org/10.1016/j.tree.2014.06.002
Zhao Z, Sun Z, Wang Z et al (2015) The high resolution sedimentary filling in Qiongdongnan Basin, Northern South China Sea. Mar Geol 361:11–24. https://doi.org/10.1016/j.margeo.2015.01.002
Yeh YC, Peres-Neto PR, Huang SW et al (2015) Determinism of bacterial metacommunity dynamics in the southern East China Sea varies depending on hydrography. Ecography 38:198–212. https://doi.org/10.1111/ecog.00986
Zhang H, Huang X, Huang L et al (2018) Microeukaryotic biogeography in the typical subtropical coastal waters with multiple environmental gradients. Sci Total Environ 635:618–628. https://doi.org/10.1016/j.scitotenv.2018.04.142
Kong J, Wang Y, Warren A et al (2019) Diversity distribution and assembly mechanisms of planktonic and benthic microeukaryote communities in intertidal zones of Southeast Fujian, China. Front Microbiol 10. https://doi.org/10.3389/fmicb.2019.02640
Zhao F, Filker S, Xu K, et al (2020) Microeukaryote communities exhibit phyla-specific distance-decay patterns and an intimate link between seawater and sediment habitats in the Western Pacific Ocean. Deep Sea Res 1 Oceanogr Res Pap 160:103279. https://doi.org/10.1016/j.dsr.2020.103279
Cordes EE, Cunha MR, Galéron J et al (2010) The influence of geological, geochemical, and biogenic habitat heterogeneity on seep biodiversity. Mar Ecol 31:51–65. https://doi.org/10.1111/j.1439-0485.2009.00334.x
Acknowledgements
We thank all members on the cruise HYDZ6-202102. We thank the crew and captain of R/V “Haiyangdizhi VI” for their great help during the work at the sea. We also thank Dr. Guangyuan Lu for his kind help on measuring environmental parameters.
Funding
This study was supported by the Hong Kong Branch of Southern Marine Science and Engineering Guangdong Laboratory (Guangzhou) (SMSEGL20SC01), the Key Special Project for Introduced Talents Team of Southern Marine Science and Engineering Guangdong Laboratory (Guangzhou) (GML2019ZD0409), and the Research Grants Council of Hong Kong (16101917 and 16101318). B.C. was supported by Southern Marine Science and Engineering Guangdong Laboratory (Guangzhou) (SMSEGL20SC02) and a Leverhulme Trust Research Project Grant (PRG-2020–389).
Author information
Authors and Affiliations
Contributions
Hongbin Liu, Zhimeng Xu, and Jiawei Chen conceived and designed the study. Jiawei Chen collected the samples and conducted the experiments. Zhimeng Xu performed the analyses and drafted the manuscript. Yingdong Li, Erfan Shekarizz, Wenxue Wu, and Bingzhang Chen gave comments and advice on the manuscript.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare no competing interests.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Xu, Z., Chen, J., Li, Y. et al. High Microeukaryotic Diversity in the Cold-Seep Sediment. Microb Ecol 86, 2003–2020 (2023). https://doi.org/10.1007/s00248-023-02212-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-023-02212-y