Abstract
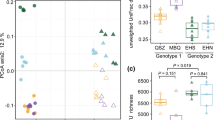
Macrophyte rhizosphere microbes, as crucial components of the wetland ecosystem, play an important role in maintaining the function and stability of natural and constructed wetlands. Distinct environmental conditions and management practices between natural and constructed wetlands would affect macrophytes rhizosphere microbial communities and their associated functions. Nevertheless, the understanding of the diversity, composition, and co-occurrence patterns of the rhizosphere bacterial communities in natural and constructed wetlands remains unclear. Here, we used 16S rRNA gene high-throughput sequencing to characterize the bacterial community of the rhizosphere and bulk sediments of macrophyte Phragmites australis in representative natural and constructed wetlands. We observed higher alpha diversity of the bacterial community in the constructed wetland than that of the natural wetland. Additionally, the similarity of bacterial community composition between rhizosphere and bulk sediments in the constructed wetland was increased compared to that of the natural wetland. We also found that plants recruit specific taxa with adaptive functions in the rhizosphere of different wetland types. Rhizosphere samples of the natural wetland significantly enriched the functional bacterial groups that mainly related to nutrient cycling and plant-growth-promoting, while those of the constructed wetland-enriched bacterial taxa with potentials for biodegradation. Co-occurrence network analysis showed that the interactions among rhizosphere bacterial taxa in the constructed wetland were more complex than those of the natural wetland. This study broadens our understanding of the distinct selection processes of the macrophytes rhizosphere-associated microbes and the co-occurrence network patterns in different wetland types. Furthermore, our findings emphasize the importance of plant–microbe interactions in wetlands and further suggest P. australis rhizosphere enriched diverse functional bacteria that might enhance the wetland performance through biodegradation, nutrient cycling, and supporting plant growth.




Similar content being viewed by others
References
Janse JH, van Dam AA, Hes EMA, de Klein JJM, Finlayson CM, Janssen ABG, van Wijk D, Mooij WM, Verhoeven JTA (2019) Towards a global model for wetlands ecosystem services. Curr Opin Env Sust 36:11–19. https://doi.org/10.1016/j.cosust.2018.09.002
Arroyo P, de Miera LES, Ansola G (2015) Influence of environmental variables on the structure and composition of soil bacterial communities in natural and constructed wetlands. Sci Total Environ 506:380–390. https://doi.org/10.1016/j.scitotenv.2014.11.039
Sundaravadivel M, Vigneswaran S (2001) Constructed wetlands for wastewater treatment. Crit Rev Environ Sci Technol 31(4):351–409. https://doi.org/10.1080/20016491089253
Vymazal J (2011) Constructed wetlands for wastewater treatment: five decades of experience. Environ Sci Technol 45(1):61–69. https://doi.org/10.1021/es101403q
Saeed T, Sun G (2012) A review on nitrogen and organics removal mechanisms in subsurface flow constructed wetlands: dependency on environmental parameters, operating conditions and supporting media. J Environ Manag 112:429–448. https://doi.org/10.1016/j.jenvman.2012.08.011
Li YF, Zhu GB, Ng WJ, Tan SK (2014) A review on removing pharmaceutical contaminants from wastewater by constructed wetlands: design, performance and mechanism. Sci Total Environ 468:908–932. https://doi.org/10.1016/j.scitotenv.2013.09.018
Faulwetter JL, Gagnon V, Sundberg C, Chazarenc F, Burr MD, Brisson J, Camper AK, Stein OR (2009) Microbial processes influencing performance of treatment wetlands: A review. Ecol Eng 35(6):987–1004. https://doi.org/10.1016/j.ecoleng.2008.12.030
Fester T, Giebler J, Wick LY, Schlosser D, Kastner M (2014) Plant-microbe interactions as drivers of ecosystem functions relevant for the biodegradation of organic contaminants. Curr Opin Biotechnol 27:168–175. https://doi.org/10.1016/j.copbio.2014.01.017
Stottmeister U, Wießner A, Kuschk P, Kappelmeyer U, Kästner M, Bederski O, Müller RA, Moormann H (2003) Effects of plants and microorganisms in constructed wetlands for wastewater treatment. Biotechnol Adv 22(1–2):93–117. https://doi.org/10.1016/j.biotechadv.2003.08.010
Philippot L, Raaijmakers JM, Lemanceau P, van der Putten WH (2013) Going back to the roots: the microbial ecology of the rhizosphere. Nat Rev Microbiol 11(11):789–799. https://doi.org/10.1038/nrmicro3109
Reinhold-Hurek B, Bünger W, Burbano CS, Sabale M, Hurek T (2015) Roots shaping their microbiome: global hotspots for microbial activity. Annu Rev Phytopathol 53(1):403–424. https://doi.org/10.1146/annurev-phyto-082712-102342
Briones AM, Okabe S, Umemiya Y, Ramsing N, Reichardt W, Okuyama H (2003) Ammonia-oxidizing bacteria on root biofilms and their possible contribution to N use efficiency of different rice cultivars. Plant Soil 250(2):335–348. https://doi.org/10.1023/A:1022897621223
Cocking EC (2003) Endophytic colonization of plant roots by nitrogen-fixing bacteria. Plant Soil 252(1):169–175. https://doi.org/10.1023/A:1024106605806
Riva V, Mapelli F, Syranidou E, Crotti E, Choukrallah R, Kalogerakis N, Borin S (2019) Root bacteria recruited by phragmites australis in constructed wetlands have the potential to enhance azo-dye phytodepuration. Microorganisms 7(10):384. https://doi.org/10.3390/microorganisms7100384
Zhao YF, Mao W, Pang LX, Li RJ, Li SQ (2020) Influence of Phragmites communis and Zizania aquatica on rhizosphere soil enzyme activity and bacterial community structure in a surface flow constructed wetland treating secondary domestic effluent in China. Environ Sci Pollut Res 27(21):26141–26152. https://doi.org/10.1007/s11356-020-08904-z
Español C, Gallardo B, Pino MR, Martín A, Comín A (2013) Is net ecosystem production higher in natural relative to constructed wetlands? Aquat Sci 75(3):385–397. https://doi.org/10.1007/s00027-012-0284-1
Zheng YC, Dzakpasu M, Wang XC, Zhang L, Ngo HH, Guo WS, Zhao YQ (2018) Molecular characterization of long-term impacts of macrophytes harvest management in constructed wetlands. Bioresour Technol 268:514–522. https://doi.org/10.1016/j.biortech.2018.08.030
Chen ZJ, Tian YH, Zhang Y, Song BR, Li HC, Chen ZH (2016) Effects of root organic exudates on rhizosphere microbes and nutrient removal in the constructed wetlands. Ecol Eng 92:243–250. https://doi.org/10.1016/j.ecoleng.2016.04.001
Pérez-Jaramillo JE, de Hollander M, Ramírez CA, Mendes R, Raaijmakers JM, Carrión VJ (2019) Deciphering rhizosphere microbiome assembly of wild and modern common bean (Phaseolus vulgaris) in native and agricultural soils from Colombia. Microbiome 7(1):114. https://doi.org/10.1186/s40168-019-0727-1
Faust K, Raes J (2012) Microbial interactions: from networks to models. Nat Rev Microbiol 10(8):538–550. https://doi.org/10.1038/nrmicro2832
Barberán A, Bates ST, Casamayor EO, Fierer N (2012) Using network analysis to explore co-occurrence patterns in soil microbial communities. ISME J 6(2):343–351. https://doi.org/10.1038/ismej.2011.119
Man Y, Wang JX, Tam NFY, Wan X, Huang WD, Tang ZY, JP, Tao R, Yang Y, (2020) Responses of rhizosphere and bulk substrate microbiome to wastewater borne sulfonamides in constructed wetlands with different plant species. Sci Total Environ 706:135955. https://doi.org/10.1016/j.scitotenv.2019.135955
Banerjee S, Schlaeppi K, van der Heijden MGA (2018) Keystone taxa as drivers of microbiome structure and functioning. Nat Rev Microbiol 16(9):567–576. https://doi.org/10.1038/s41579-018-0024-1
Srivastava J, Kalra SJS, Naraian R (2014) Environmental perspectives of Phragmites australis (Cav.) Trin. Ex Steudel Appl Water Sci 4:193–202. https://doi.org/10.1007/s13201-013-0142-x
Tercero MC, Álvarez-Rogel J, Conesa HM, Ferrer MA, Calderón AA, López-Orenes A, González-Alcaraz MN (2015) Response of biogeochemical processes of the water-soil-plant system to experimental flooding-drying conditions in a eutrophic wetland: the role of Phragmites australis. Plant Soil 396(1–2):109–125. https://doi.org/10.1007/s11104-015-2589-z
İnceoğlu Ö, Salles JF, van Overbeek L, van Elsas JD (2010) Effects of plant genotype and growth stage on the betaproteobacterial communities associated with different potato cultivars in two fields. Appl Environ Microbiol 76(11):3675–3684. https://doi.org/10.1128/AEM.00040-10
Zeng J, Jiao CC, Zhao DY, Xu HM, Huang R, Cao XY, Yu ZB, Wu QLL (2019) Patterns and assembly processes of planktonic and sedimentary bacterial community differ along a trophic gradient in freshwater lakes. Ecol Indic 106:105491. https://doi.org/10.1016/j.ecolind.2019.105491
Hu SW, He RJ, Wang WJ, Zhao DY, Zeng J, Huang R, Duan M, Yu ZB (2021) Composition and co-occurrence patterns of Phragmites australis rhizosphere bacterial community. Aquat Ecol 55:695–710. https://doi.org/10.1007/s10452-021-09855-4
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Tumbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R (2010) QIIME allows analysis of high-throughput communitysequencing data. Nat Methods 7(5):335–336. https://doi.org/10.1038/nmeth.f.303
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120. https://doi.org/10.1093/bioinformatics/btu170
Magoč T, Salzberg SL (2011) FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27(21):2957–2963. https://doi.org/10.1093/bioinformatics/btr507
Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27(16):2194–2200. https://doi.org/10.1093/bioinformatics/btr381
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naїve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73(16):5261–5267. https://doi.org/10.1128/AEM.00062-07
Price MN, Dehal PS, Arkin AP (2009) FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol 26(7):1641–1650. https://doi.org/10.1093/molbev/msp077
Posada D, Buckley TR (2004) Model selection and model averaging in phylogenetics: advantages of Akaike information criterion and Bayesian approaches over likelihood ratio tests. Syst Biol 53:793–808. https://doi.org/10.1080/10635150490522304
Faust K, Sathirapongsasuti JF, Izard J, Segata N, Gevers D, Raes J, Huttenhower C (2012) Microbial co-occurrence relationships in the human microbiome. PLoS Comput Biol 8(7):e1002606. https://doi.org/10.1371/journal.pcbi.1002606
Bastian M, Heymann S, Jacomy M (2009) Gephi: an open source software for exploring and manipulating networks. In: International AAAI Conference on Weblogs and Social Media. San Jose; https://gephi.org/users/publications/
Newman MEJ (2006) Modularity and community structure in networks. Proc Natl Acad Sci U S A 103(23):8577–8582. https://doi.org/10.1073/pnas.0601602103
Guimera R, Amaral LAN (2005) Functional cartography of complex metabolic networks. Nature 433(7028):895–900. https://doi.org/10.1038/nature03288
Ansola G, Arroyo P, de Miera LES (2014) Characterisation of the soil bacterial community structure and composition of natural and constructed wetlands. Sci Total Environ 473:63–71. https://doi.org/10.1016/j.scitotenv.2013.11.125
He RJ, Zeng J, Zhao DY, Wang SR, Wu QLL (2021) Decreased spatial variation and deterministic processes of bacterial community assembly in the rhizosphere of Phragmites australis across the Middle-Lower Yangtze plain. Mol Ecol. https://doi.org/10.1111/mec.16298
Huang R, Zeng J, Zhao DY, Cook KV, Hambright KD, Yu ZB (2020) Sediment microbiomes associated with the rhizosphere of emergent macrophytes in a shallow, subtropical lake. Limnol Oceanogr 65:S38–S48. https://doi.org/10.1002/lno.11325
He RJ, Zeng J, Zhao DY, Huang R, Yu ZB, Wu QLL (2020) Contrasting patterns in diversity and community assembly of phragmites australis root-associated bacterial communities from different seasons. Appl Environ Microbiol 86(14):e00379-e420. https://doi.org/10.1128/AEM.00379-20
Wu FY, Chung AKC, Tam NFY, Wong MH (2012) Root exudates of wetland plants influenced by nutrient status and types of plant cultivation. Int J Phytorem 14(6):543–553. https://doi.org/10.1080/15226514.2011.604691
Luo P, Liu F, Zhang SN, Li HF, Yao R, Jiang QW, Xiao RL, Wu JS (2018) Nitrogen removal and recovery from lagoon-pretreated swine wastewater by constructed wetlands under sustainable plant harvesting management. Bioresour Technol 258:247–254. https://doi.org/10.1016/j.biortech.2018.03.017
Cao QQ, Wang H, Chen XC, Wang RQ, Liu J (2017) Composition and distribution of microbial communities in natural river wetlands and corresponding constructed wetlands. Ecol Eng 98:40–48. https://doi.org/10.1016/j.ecoleng.2016.10.063
Zhou JZ, Deng Y, Zhang P, Liang YT, Van Nostrand JD, Yang YF, He ZL, Wu LY, Stahl DA, Hazen TC, Tiedje JM, Arkin AP (2014) Stochasticity, succession, and environmental perturbations in a fluidic ecosystem. Proc Natl Acad Sci U S A 111(9):E836–E845. https://doi.org/10.1073/pnas.1324044111
Zuo XJ, Zhang HS, Yu JH (2020) Microbial diversity for the improvement of nitrogen removal in stormwater bioretention cells with three aquatic plants. Chemosphere 244:125626. https://doi.org/10.1016/j.chemosphere.2019.125626
Zhu Y, Zhang X, Wu X, Chen G, Bakken LR, Zhao L, Frostegard A, Zhang X (2017) Carbon-driven enrichment of the crucial nitrate-reducing bacteria in limed peat soil microcosms. Lett Appl Microbiol 65(2):159–164. https://doi.org/10.1111/lam.12756
Deng M, Li L, Dai ZL, Senbati Y, Song K, He XG (2020) Aerobic denitrification affects gaseous nitrogen loss in biofloc-based recirculating aquaculture system. Aquaculture 529:735686. https://doi.org/10.1016/j.aquaculture.2020.735686
Li M, Duan R, Hao W, Li QC, Arslan M, Liu PP, Qi X, Huang X, El-Din MG, Liang P (2020) High-rate nitrogen removal from carbon limited wastewater using sulfur-based constructed wetland: impact of sulfur sources. Sci Total Environ 744:1140969. https://doi.org/10.1016/j.scitotenv.2020.140969
Shameer S, Prasad T (2018) Plant growth promoting rhizobacteria for sustainable agricultural practices with special reference to biotic and abiotic stresses. Plant Growth Regul 84(3):603–615. https://doi.org/10.1007/s10725-017-0365-1
Haney CH, Samuel BS, Bush J, Ausubel FM (2015) Associations with rhizosphere bacteria can confer an adaptive advantage to plants. Nat Plants 1(6):1–9. https://doi.org/10.1038/NPLANTS.2015.51
Kasanke CP, Collins RE, Leigh MB (2019) Identification and characterization of a dominant sulfolane-degrading Rhodoferax sp via stable isotope probing combined with metagenomics. Sci Rep 9:3121. https://doi.org/10.1038/s41598-019-40000-2
Sun WM, Xie SG, Luo CL, Cupples AM (2010) Direct link between toluene degradation in contaminated-site microcosms and a Polaromonas strain. Appl Environ Microbiol 76(3):956–959. https://doi.org/10.1128/AEM.01364-09
Jechalke S, Franchini AG, Bastida F, Bombach P, Rosell M, Seifert J, von Bergen M, Vogt C, Richnow HH (2013) Analysis of structure, function, and activity of a benzene-degrading microbial community. FEMS Microbiol Ecol 85(1):14–26. https://doi.org/10.1111/1574-6941.12090
Sun BZ, Ko K, Ramsay JA (2011) Biodegradation of 1,4-dioxane by a Flavobacterium. Biodegradation 22(3):651–659. https://doi.org/10.1007/s10532-010-9438-9
Tonolla M, Demarta A, Peduzzi S, Hahn D, Peduzzi R (2000) In situ analysis of sulfate-reducing bacteria related to Desulfocapsa thiozymogenes in the chemocline of meromictic Lake Cadagno (Switzerland). Appl Environ Microbiol 66(2):820–824. https://doi.org/10.1128/AEM.66.2.820-824.2000
El Houari A, Ranchou-Peyruse M, Ranchou-Peyruse A, Dakdaki A, Guignard M, Idouhammou L, Bennisse R, Bouterfass R, Guyoneaud R, Qatibi AI (2017) Desulfobulbus oligotrophicus sp nov, a sulfate reducing and propionate oxidizing bacterium isolated from a municipal anaerobic sewage sludge digester. Int J Syst Evol Microbiol 67(2):275–281. https://doi.org/10.1099/ijsem.0.001615
Eshinimaev BT, Medvedkova KA, Khmelenina VN, Suzina NE, Osipov GA, Lysenko AM, Trotsenko YA (2004) New thermophilic methanotrophs of the genus Methylocaldum. Microbiology 73(4):448–456. https://doi.org/10.1023/B:MICI.0000036991.31677.13
Hubbell SP (2005) Neutral theory in community ecology and the hypothesis of functional equivalence. Funct Ecol 19(1):166–172. https://doi.org/10.1111/j.0269-8463.2005.00965.x
Zhang SQ, Huang Y, Xing JL, Chen ZJ, Meng FG (2020) Response of anammox metacommunity to varying hydrodynamic wash. J Water Process Eng 33:101096. https://doi.org/10.1016/j.jwpe.2019.101096
Ahsan M, Riaz A, Jaskani MJ, Hameed M (2017) Physiological and anatomical response of fragrant rosa species with treated and untreated wastewater. Int J Agric Biol 19(1):13–22. https://doi.org/10.17957/IJAB/15.0160
Hassani MA, Durán P, Hacquard S (2018) Microbial interactions within the plant holobiont. Microbiome 6:58. https://doi.org/10.1186/s40168-018-0445-0
de Vries FT, Griffiths RI, Bailey M, Craig H, Girlanda M, Gweon HS, Hallin S, Kaisermann A, Keith AM, Kretzschmar M, Lemanceau P, Lumini E, Mason KE, Oliver A, Ostle N, Prosser JI, Thion C, Thomson B, Bardgett RD (2018) Soil bacterial networks are less stable under drought than fungal networks. Nat Commun 9:3033. https://doi.org/10.1038/s41467-018-05516-7
Coyte KZ, Schluter J, Foster KR (2015) The ecology of the microbiome: networks, competition, and stability. Science 350:663–666. https://doi.org/10.1126/science.aad2602
Xue YF, Tian J, Quine TA, Powlson D, Xing KX, Yang LY, Kuzyakov Y, Dungait JAJ (2020) The persistence of bacterial diversity and ecosystem multifunctionality along a disturbance intensity gradient in karst soil. Sci Total Environ 748:142381. https://doi.org/10.1016/j.scitotenv.2020.142381
Deng Y, Jiang YH, Yang YF, He ZL, Luo F, Zhou JZ (2012) Molecular ecology network analyses. BMC Bioinf 13:113. https://doi.org/10.1186/1471-2105-13-113
Zhou JZ, Deng Y, Luo F, He ZL, Tu QC, Zhi XY (2010) Functional molecular ecological networks. mBio 1(4):e00169-e210. https://doi.org/10.1128/mBio.00169-10
Xue L, Ren HD, Li S, Leng XH, Yao XH (2017) Soil bacterial community structure and co-occurrence pattern during vegetation restoration in karst rocky desertification area. Front Microbiol 8:2377. https://doi.org/10.3389/fmicb.2017.02377
Wang LX, Han MZ, Li X, Yu BB, Wang HD, Ginawi A, Ning K, Yan YJ (2021) Mechanisms of niche-neutrality balancing can drive the assembling of microbial community. Mol Ecol 30(6):1492–1504. https://doi.org/10.1111/mec.1582578
Hartwell LH, Hopfield JJ, Leibler S, Murray AW (1999) From molecular to modular cell biology. Nature 402(6761):C47–C52. https://doi.org/10.1038/35011540
Chee-Sanford J, Tian D, Sanford R (2019) Consumption of N2O and other N-cycle intermediates by Gemmatimonas aurantiaca strain T-27. Microbiology-Sgm 165(12):1345–1354. https://doi.org/10.1099/mic.0.000847
Yang B, Yin F, Wang CM, Zhao XL, Liu J, Wu K, Yang L, Zhang WD (2019) Construction of biogas metabolic pathway in a low-temperature biogas fermentation system. Energy Sci Eng 7(6):3160–3173. https://doi.org/10.1002/ese3.488z
Bukowska A, Kalinski T, Chrost RJ (2019) Degradation of microcystins by water and bottom sediment bacterial communities from a eutrophic freshwater lake. Aquat Microb Ecol 82(2):129–144. https://doi.org/10.3354/ame01887
Funding
This work was supported by the National Natural Science Foundation of China (32022050, 31730013, U2040201, 31971478 and 32171563), the Second Tibetan Plateau Scientific Expedition and Research Program (2019QZKK0503), Key Research Program of Frontier Science, CAS (QYZDJ-SSW-DQC030), and Project of Young Scientist Group of NIGLAS (2021NIGLAS-CJH01).
Author information
Authors and Affiliations
Contributions
Siwen Hu: data curation, writing—original draft preparation, formal analysis, visualization. Rujia He: investigation, formal analysis, writing—reviewing, and editing. Jin Zeng: conceptualization, writing—reviewing, and editing, supervision, funding acquisition. Dayong Zhao: conceptualization, methodology, resources, writing—reviewing and editing, funding acquisition, supervision. Shuren Wang: investigation, experiment. Fei He: writing—reviewing and editing. Zhongbo Yu: conceptualization, writing—reviewing, and editing. Qinglong L. Wu: conceptualization, funding acquisition.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare no conflict of interest.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Hu, S., He, R., Zeng, J. et al. Lower Compositional Variation and Higher Network Complexity of Rhizosphere Bacterial Community in Constructed Wetland Compared to Natural Wetland. Microb Ecol 85, 965–979 (2023). https://doi.org/10.1007/s00248-022-02040-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-022-02040-6