Abstract
Hematophagous Spinturnix myoti mites and their host, the greater mouse-eared bat (Myotis myotis), were tested for the presence of Bartonella spp., Rickettsia spp., and Anaplasma phagocytophilum. In total, Bartonella spp. DNA was amplified in 28% of 134 mite pools and in 25% of 59 bats tested by PCR targeting a fragment of citrate synthase gltA gen. Adult mites were at least threefold more frequently infected compared to immature stages. The overall infection prevalence among mite pools from cave-dwelling bats was higher than for those collected from attic shelters. Three distinct genotypes were detected. The most prevalent genotype in mites and bats matched closely with Candidatus Bartonella hemsundetiensis identified in bats from Finland and was relatively distant from bat-borne Bartonella strains described in the UK and France. Importantly, most sequences were close to those reported in forest workers from Poland. The presence of identical genotype among S. myoti samples and M. myotis bats suggests that bartonellae can be shared between mites and their bat hosts. In this case, wing mites could serve as vectors, whereas their hosts as reservoirs. One blood sample was positive by PCR for the msp2 gene of A. phagocytophilum. Two mite pools yielded Rickettsia spp. DNA. Both sequences were distinct from any known species but can be classified as spotted fever group Rickettsia spp. Our findings expanded our knowledge on the role of spinturnicid mites in the ecology and epidemiology of bacterial infections associated with vespertilionid bats, especially regarding the genus Bartonella.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In the ecology and epidemiology of vector-borne zoonotic diseases, it is essential to identify (i) a group of vertebrate reservoir species acting as a natural source for a pathogen and (ii) competent biological vectors which ensure its multiplication, long-term maintenance, and finally active transmission to hosts. These components are responsible for emergence and maintenance of endemic foci of vector-borne diseases. Bats from the order Chiroptera are highly mobile flying vertebrates which comprise one fourth of the world’s species of mammals and frequently host heavy loads of highly specialized groups of hematophagous arthropods. Bat-adapted flies (Nycteribiidae, Streblidae), fleas (Ischnopsyllidae), bugs (Cimicidae), mites (e.g., Macronyssidae, Spinturnicidae), and ticks (Ixodidae and Argasidae) may carry various zoonotic pathogens and probably act as their vectors within chiropteran populations [1].
Mites of the family Spinturnicidae (Acari, Mesostigmata) belong to the most abundantly and regularly recorded non-tick acarines associated with bats from the suborder Yangochiroptera and with two families (Rhinopomatidae, Rhinolophidae) from the suborder Yinpterochiroptera, As permanent ectoparasites (without free-living stages) exhibit high host specificity completing their entire life cycle on one or a few closely related bat species [2, 3], they infest only bare fragments of bat’s skin like wing and tail membranes and therefore are called wing mites. Their reproduction period is strictly synchronized with the breeding cycle of the host. Consequently, the highest numbers of mites with prevalence values up to 100% are recorded on pregnant and lactating females, and especially on their offspring, which in Europe are nursed for several summer weeks (from June to July) in colonial roosts [3,4,5]. Whereas eggs and larval stages develop inside a pregnant female mite, each of active mobile developmental stages (the protonymph, deutonymph, and adult) feeds several times on the host’s blood and lymph [6]. To date, in Europe, the family Spinturnicidae comprises 15 species [7] and 13 of them have been confirmed on bats in Poland [8].
Repeated feedings and frequent co-infestation of European bats by bat-associated tick species, such as Argas vespertilionis, Ixodes vespertilionis, I. ariadnae, and occasionally I. ricinus, strongly imply that these mites are naturally exposed to various blood-borne bacterial agents [9,10,11]. However, the potential role of Spinturnicidae as biological vectors, reservoirs, or amplifiers for bacterial agents circulating in bat communities remains still poorly studied. There are only two published reports on bacterial agents detected in wing mites. Revees et al. [12] confirmed the presence of the 16S rRNA and p44 genes identical to Anaplasma phagocytophilum in Spinturnix psi collected from a bat of the genus Miniopterus in Madagascar. The sequence of the p44 gene showed 100% identity to those found in human and horse isolates with granulocytic anaplasmosis. Furthermore, Hornok et al. [9] identified Bartonella spp. DNA in Spinturnix myoti pools obtained from three Myotis myotis bats in Hungary.
The aim of our work was to investigate the presence of potentially zoonotic agents such as Bartonella spp., Rickettsia spp., and Anaplasma phagocytophilum in Spinturnix myoti mites and their main host, the greater mouse-eared bat, Myotis myotis, by PCR assays. These intracellular bacteria are predominantly transmitted by hematophagous arthropod vectors including mites and ticks [1]. The mode of transmission of these agents among populations of European bat species as well as vector competency of their highly specialized ectoparasites remains unknown. The greater mouse-eared bat is one of the largest European bats from the insectivorous family Vespertilionidae. During its reproduction period, it resides in caves or in large buildings, including church or school attics where it forms breeding aggregations, and therefore besides Eptesicus serotinus, it is recognized as the most common synanthropic species [13, 14]. It is parasitized with the Spinturnix myoti mite which may also occur on a few other Myotis species and only sporadically on bats of genera Barbastella, Pipistrellus, Plecotus, and Vespertilio [14]. This wing mite has a wide distribution range in Europe and additionally has been recorded in North Africa and Asia [15]. In the present study, bats were sampled for peripheral blood and mites in cave and attic roosting shelters localized in west-central and southern Poland.
Materials and Methods
Study Sites
Bats were sampled in five shelters of Myotis myotis located in west-central and southern Poland in years 2007–2008 (Fig. 1). The chosen shelters were represented by two caves: (1) Studnisko cave (Krakowsko-Wieluńska Upland, 346 m a.s.l., 19° 16′ E; 50° 43′ N) and (2) Szachownica cave (Śląsko-Krakowska Upland, 215 m a.s.l., 18° 48′ E; 51° 03′ N), and three attics: (3) a school attic in Kopanki (Wielkopolska Province, west-central Poland, 108 m a.s.l., 16° 18′ E, 52° 17′ N) and two church attics situated in (4) Skalnik and (5) Nowosielce (the Beskid Mountains, a part of the Carpathians: 350 m a.s.l., 21° 28′ E, 49° 34′ N, and 330 m a.s.l., 22° 23′ E, 50° 03′ N, respectively).
Bat Capture and Mite Collection
In this study, only bats infested with spinturnicid mites were analyzed. Most of them (72 out of 80) were captured during their breeding season (from mid-May to mid-August) by hand directly from aggregations in attics or using mist nets and harp traps at colony entrances of caves. Eight individuals from the Szachownica cave were mist-netted during their mating period in October. Bats were placed into clean separate bags and identified based on differences in their morphological characteristics [16]. Sex and age (adult, juvenile) were determined for each caught individual. EDTA-blood samples (approximately 5 μL) were successfully collected from 59 of 80 (73.7%) bats by a puncture of the peripheral uropatagium vein using a 28-gauge needle and a pipette (Table 2). A fast acting gel (Super Clot Gel, Synergy Labs) was applied to the puncture site to disinfect it and avoid bleeding. Blood samples were stored at − 20 °C in Eppendorf tubes until molecular surveys. Spinturnicid mites were collected from wing and tail membranes of their hosts using forceps and preserved in 75% ethanol for further analyses. Their morphological identification was based on the keys by Dusbábek [17].
After collection of mites and blood, each animal was released in its natural habitat. Trapping and handling procedures of bats were approved by the permission of the Polish Ministry of Environment (no. DLOPiK-op/Ozgi-4200/IV.D-20/7666/06/aj). Due to the low number of adult males (n = 4), only adult females and juvenile bats were analyzed for differences in mean intensity calculated as mean number of mites per infested host ± standard deviation (SD) during the breeding season. The eight animals from the Szachownica cave, which were mist-netted in the postbreeding period in October, were excluded from the statistical analysis regarding infestation parameters.
DNA Extraction, PCR Assays, and Sequencing
DNA was extracted from blood samples of bats and from spinturnicid mites by use of the commercial kits: Genomic Mini AX Blood and Sherlock AX (A&A Biotechnology, Gdynia, Poland), respectively. Mites were processed in pools containing between two and ten individuals derived from the same bat. Two groups of mites: (i) immature stages (protonymphs and deutonymphs) and (ii) adult stages were polled and tested separately. A total of 130 DNA samples was obtained from spinturnicid mites.
Samples were analyzed for the presence of Bartonella spp., Rickettsia spp., and A. phagocytophilum by conventional PCR assays. Bartonella spp. were detected using primers with primers BhCS781 and BhCS1137 targeting the citrate synthase gltA gen fragment with expected size 379 bp and under the cycling conditions as previously described [18]. Rickettsial DNA was detected using the RpCs.877 and RpCs.1258 primers amplifying a 381-bp fragment of the gltA gene which has conserved regions shared by all known Rickettsia species [19]. A species-specific direct PCR assay that amplifies a 334-bp fragment of the msp2 gene (primers: MSP2-3f and MSP2-3r) was used for the detection of A. phagocytophilum [20, 21]. To determine genetic variants of A. phagocytophilum, a nested PCR amplifying a 546-bp region of the 16S rRNA gene was used as previously reported [22]. Positive and negative (doubled distilled water) controls were included with each PCR. PCR products were separated by 1.5% agarose gel electrophoresis and visualized by ethidium bromide. Selected PCR-positive products were sequenced in both directions using PCR primers. Sequencing products were resolved by using an ABI 3100 automated sequencer (PerkinElmer). Sequence analysis was performed by using the software package ABI Prism DNA Sequencing Analysis Software version 3.0 (PerkinElmer). All obtained sequences were compared with those available in GenBank databases using BLAST (www.ncbi.nlm.nih.gov/blast/Blast.cgi). Phylogenetic dendrograms with Bartonella spp. and Rickettsia spp. gltA sequences obtained in this study and selected sequences deposited in GenBank were constructed by the neighbor-joining algorithm method using CLC Sequence Viewer version 7.6. Jukes-Cantor model was used for nucleotide distance measurement. Representative partial sequences determined in this study were deposited in GenBank under the following accession numbers: JQ695834-40 for the gltA gene of Bartonella spp. and JQ695832 and JQ695833 for the gltA gene of Rickettsia spp. Rates of infection were analyzed by use of chi-squared test χ2, whereas differences in mean intensity of mite infestation by Mann-Whitney U test with P < 0.05 were considered statistically significant.
Results
Altogether, 80 individuals of the greater mouse-eared bat were included in this study: 43 adult females, 19 young females, four adult males, and 14 young males. A total of 707 spinturnicid mites, including 400 adults (56.6%) and 307 immature individuals (43.4%), were removed from the captured hosts (Table 1). All were identified as Spinturnix myoti Kolenati 1856.
Intensity of Mite Infestation
Mean (± SD, range) mite intensities assessed for adult females (n = 43) and juvenile bats (including 15 females and 11 males) during the breeding season were similar (9.8 ± 4.6 and 8.9 ± 4.0, respectively). Cave-dwelling bats hosted higher mite loads than those from attics (10.9 ± 3.9 vs. 7.5 ± 4.0 per bat, respectively, Mann–Whitney U test p = 0.0004).
Bacterial Infections in Mites
DNA of Bartonella spp. was amplified in 38 (28.4%) of the 134 S. myoti pools (Table 1). The positive samples were collected from 35 (43.7%) of the 80 bats, including 23 (48.9%) out of 47 adults and 12 (36.4%) out of 33 juveniles. Adult mites were at least threefold more frequently infected with the bacterium compared to immature stages (40.3 and 12.3%, respectively, χ2 test P = 0.0003). The overall infection prevalence among mite pools removed from cave-dwelling bats was significantly higher than in those collected from bats in attic shelters (34.4 and 15.9%, respectively; χ2 test P = 0.025). Animals infested with at least one PCR positive pool were also more frequent in the former group in comparison with bats from the latter (56.0 vs. 23.3%; χ2 test P = 0.004). Three animals from Studnisko cave were concurrently infested with infected adult and immature stages of S. myoti.
Three novel gltA gene sequences of Bartonella spp. termed as genotypes A, B, and C were successfully generated from representative PCR positive mite pools (Table 3). Their nucleotide identity to each other ranged from 96.3 to 99.7%. The genotype A proved to be the most common and was identified in 18 (90%) out of 20 PCR amplicons, whereas the variants B and C were detected only in two single pools. The dominant genotype A shared 100% (299/299 bp) identity with Bartonella strains isolated from three bat species: Myotis blythii (GenBank e.g., KX300136), M. emarginatus (isolate B44736), and Eptesicus serotinus (isolate B44714) sampled in Georgia. Furthermore, the genotypes A and B exhibited 100 and 99.4% similarity to human-derived sequences identified in forest workers in Poland (e.g., GenBank HM116786). Both variants were also closely related to the recently reported Candidatus Bartonella hemsundetiensis (GenBank KR822802) from M. daubentonii bats captured in Finland (mean sequence similarity = 99.5%). The variant C reached 96.3% nucleotide identity. Our samples were distinct from Bartonella spp. described from bats in the UK and France. Figure 2 shows the phylogenetic relationships of the Bartonella spp. gltA genetic variants detected in mites and bats in this study with selected sequences available on GenBank.
Phylogenetic relationships of Bartonella spp. genotypes found in Spinturnix myoti mites and Myotis myotis bats based on the fragment of gltA gene of Bartonella spp. and selected sequences available from GenBank. Inference was made by using the neighbor joining method in CLC Sequence Viewer Version 7.6. Jukes-Cantor model was used for nucleotide distance measurement. Bootstrap analysis with 100 replicates was performed. The scale represents 0.03 substitution per base per indicated horizontal distance. Note: branches shorter than 0.0022 are shown as having length 0.0022. The gapped branches have been truncated to one third of their original length for clarity. The tree is rooted with gltA gene of B. tamiae [DQ395177]. Bartonella spp. genotypes found in blood of bats and in mite pools in this study were marked with one and two asterisks, respectively
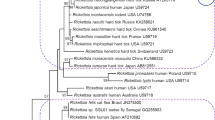
Rickettsia spp. DNA was detected in two (1.5%) pools comprising adult mites (Table 1). Sequence analysis of both positive products with gltA-specific primers revealed that they were identical to one another and unique from other known Rickettsia spp., with DNA similarity values < 92% (GenBank acc. nos. JQ695832, JQ695833). They showed maximum similarity score of 91.8% (312/340 bp) with R. massiliae (KJ663740) and Candidatus R. barbariae (EU272185) detected in Rhipicephalus sanguineus and R. turanicus ticks, respectively. Moreover, both samples shared 91.5% similarity with the Rickettsia sp. AvBat strain isolated from Argas vespertilionis in France (JN038177), and 91.2% similarity with Candidatus Rickettsia wissemanii identified in Ornithodoros hasei in French Guiana (LT558852) and Candidatus Rickettsia nicoyana isolated from Ornithodoros knoxjonesi in Costa Rica (KX228143). Bats are hosts for the mentioned ticks. The gltA phylogenetic tree reflected these relationships with a distinct position of both our sequences which clustered in a separate clade (Fig. 3). All mite samples tested negative for DNA of A. phagocytophilum by using primers targeting the msp2 gene.
Phylogenetic relationships of the two Rickettsia genotypes found in Spinturnix myoti mites species based on the fragment of gltA gene of Rickettsia spp. and selected sequences available from GenBank. Inference was made by using the neighbor joining method in CLC Sequence Viewer Version 7.6. Bootstrap analysis with 100 replicates was performed. The scale represents 0.015 substitution per base per indicated horizontal distance. Note: branches shorter than 0.0015 are shown as having length 0.0015. The gapped branches have been shortened by 0.03 for clarity. The tree is rooted with gltA gene of R. bellii [DQ146481]. Selected Rickettsia spp. sequences from bat-associated ticks and several bat species were marked with one and two asterisks, respectively
Bacterial Infections in Bats
Bartonella spp. DNA was detected in 15 (25.4%) of the 59 blood samples, with the range from 12.5 to 29.7% depending on the collection site (Table 2). Blood PCR positive animals were recorded both in cave and in school attic shelters. The infection prevalence was comparable among juvenile and adult bats (22.2 and 28.1%, respectively). Of the 15 bacteremic animals, 11 (73%) carried also PCR-positive mite pools. Moreover, two of the 11 bats hosted concurrently infected adult and immature S. myoti stages. Sequence analysis of 10 selected gltA gene sequences identified in the bats demonstrated 100% identity to the genotype A found in most infected mite pools (Table 3). Two representatives of the gltA sequences from M. myotis were submitted to the GenBank (JQ695834, JQ695835).
Rickettsia spp. DNA was not detected in any of the 59 blood samples. One female, derived from Kopanki school attic, yielded a 334-bp fragment of the A. phagocytophilum msp2 gene. Unfortunately, due to a limited volume of DNA extract from this single sample, a confirmation of the amplified A. phagocytophilum DNA by the 16S rRNA gene nested PCR assay and subsequent sequencing was unsuccessful.
Discussion
In this report, we screened using PCR assays hematophagous Spinturnix myoti mites and their natural host, the greater mouse-eared bat, Myotis myotis, for the presence of medically important arthropod-borne intracellular bacteria: Bartonella spp., Rickettsia spp., and A. phagocytophilum. Among these agents, bartonellae proved to be the most common infection both in mites and their hosts. Overall, 28% S. myoti pools and 25% M. myotis blood samples yielded these bacteria. The presence of bartonellae in S. myoti mites was previously reported in a survey conducted in Hungary, in which each of the three pools tested (comprising 85 mites) by TaqMan PCR yielded Bartonella spp. [9].
The infection prevalence in mites sampled in our study varied depending on a developmental stage and a shelter type of their hosts. Pools with adult mites harbored the bacterium at least threefold more frequently than those with immature stages (40 vs. 12%). Given that spinturnicid mites feed multiple on an individual host for at least several months, these differences could be attributed to previous episodes of the adult mites which would have fed upon bacteriemic animals. In our opinion, detection of Bartonella DNA both in adult and subadult S. myoti pools may imply that spinturnicid mites are not only exposed to the infection but they might also maintain it in subsequent developmental stages (transstadial transmission). The presence of infected pools with adult mites collected from 18 blood-negative bats seems to support this supposition, especially that none of 13 immature mite pools, obtained from the same hosts, yielded the bacterium (data not shown). Bartonella species invade erythrocytes and endothelial cells causing usually long-lasting infections accompanied by a relapsing intraerythrocytic bacteremia of variable level which in their mammalian reservoir hosts can persist months or even years [23]. Therefore, spinturnicid mites, as permanent ectoparasites with multiple feeding mode observed in each life stage, have ample opportunities to encounter and acquire blood-borne pathogens during a bacteremic period of the host. Furthermore, their typically high abundance and prevalence up to 100% in summer maternity colonies [24], vertical (from mother to offspring) and horizontal (among individuals) transfer between tightly roosting animals, clearly increase their dispersal capacity and make them at least theoretically ideal vectors for various blood-borne bacterial agents. Of note is that the mean mite intensity values recorded on adult females and juvenile bats during the breeding season were comparable (10.6 and 8.9 mites per bat, respectively). This finding confirms previous results (e.g., [3, 4]) and demonstrates a high dispersion potential of wing mites which may facilitate transfer of blood-borne bacteria among bats.
Interestingly, mite samples from cave dwelling bats were twice more frequently infected than those collected from attic-dwelling hosts (34 vs. 16%). Similarly, in a study conducted by Sándor et al. [25], the highest prevalence of Bartonella spp. infected bat flies (Nycteribiidae) was recorded among bat species (Rhinolophus euryale, Myotis capaccinii, Miniopterus schreibersii) roosting exclusively in caves. Moreover, in our study, a higher proportion of cave-dwelling animals hosted PCR positive pools in comparison with those sampled in attics (56 vs. 27%). This indicates that bats roosting in caves colonies were more often exposed to infected mites that could be attributed to higher S. myoti loads recorded on animals breeding in this type of shelter. Although M. myotis can breed successfully in micro-climatically distinct nursery roosts [26], conditions prevailing in caves with year-round stable microclimate seem to be particularly favorable for a larger reproduction of spinturnicid mites and may explain differences in infestation parameters observed in our study. Postawa et al. [24] demonstrated that greater mouse-eared bats from cave nursery colonies harbored at least threefold more S. myoti mites than those from attic colonies, irrespective of host sex or age. The authors concluded that the microclimate of the host’s roosts favor ectoparasite abundance. In our opinion, higher loads of spinturnicid mites especially on bacteremic animals may not only increase the effectiveness of their horizontal and vertical transfer but might also contribute to higher prevalence of Bartonella spp. in S. myoti mites collected from cave-dwelling bats as we observed in the present study. Moreover, they may act as amplifiers for horizontal transmission of bat-associated Bartonella spp. to competent vectors (e.g., bat bugs and argasid ticks) which are known to attack occasionally humans [1, 9].
The detection of Bartonella spp. DNA in peripheral blood of 25% greater mouse-eared bats sampled in our study demonstrates their susceptibility to bartonellae infections which seem to be rather frequent in populations of this bat species. Comparison of available data regarding Bartonella infection among European bats from the family Vespertilionidae showed that the mean prevalence found in M. myotis was higher than the 8.8% reported in bats from the UK or the 10% described in France, but lower than that of 37% in Finland, and 38.7% in Georgia, a country in the Caucasus region located at the border of Eastern Europe [27,28,29,30]. These bat-borne Bartonella spp. have been found so far in ten vespertilionid bat species. Five of them belong to the genus Myotis: M. blythii, M. daubentonii, M. emarginatus, M. mystacinus, and M. myotis (this study). The remaining five species comprise: Eptesicus nilssoni, E. serotinus, Pipistrellus nathusii, P. pygmaeus, and Nyctalus noctula. Interestingly, in our study, the majority of blood-positive bats (73%) hosted concurrently infected mite pools, which implies that these mites might have acquired the bacterium during feeding upon bacteremic hosts. This indicates that M. myotis may develop active bacteremia and maintain the circulation of bartonellae acting as a competent reservoir. In our opinion, co-occurrence of infected adult and immature stages of S. myoti on two PCR-positive bats, additionally, supports this presumption. On the other hand, the finding that juvenile bats and adult females (excluding animals from Szachownica) showed identical infection rates (25%) suggests a potential role of spinturnicid mites in active acquisition and transmission of Bartonella spp. between mothers and their offspring during the reproductive season. This hypothesis appears to be strongly supported by the fact that PCR-positive M. myotis bats and infected mite pools yielded the same genotype A of the bacterium.
Analysis of the partial gltA gene sequences of Bartonella spp. found in S. myoti pools revealed the presence of three genotypes A, B, and C, with evident predominance of the former. Remarkably, the genotype A proved to be the only variant identified among bacteriemic M. myotis bats. The genotype A exhibited 100% similarity to Bartonella strains recently isolated from three bat species sampled in Georgia representing the largest genogroup Vesp-6 dominated by M. blythii [30]. Unexpectedly, all of the Bartonella genotypes identified in our study were phylogenetically distant from those found in bats from UK and France [27, 29]. They were also distant from strains reported from Finland [31] including B. naantaliensis and two strains detected in M. daubentonii and E. nilssoni closely resembling Candidatus B. mayotimonensis isolated from an endocarditis human patient in the USA [32]. On the other hand, the genotypes A and B in our study, as well as Georgian strains from Vesp-6 genogroup, clustered along with Candidatus B. hemsundetiensis recently detected by Lilley et al. [28] in blood of M. daubentonii bats from Finland. In our opinion, postglacial recolonization of Europe by several Myotis spp. from the region of Caucasus [33] might contribute to the observed phylogenetic similarity of Bartonella sequences among samples from M. myotis representing populations of Central Europe and Georgian bats from Ves-6 genogroup belonging to Caucasian populations.
It is noteworthy that the genotypes A and B were genetically similar to sequences previously detected in forest workers in Poland, however with unknown pathogenicity [34]. Although some bat-associated bartonellae described in the UK, Finland, and France resemble Candidatus B. mayotimonensis, an etiological agent of human endocarditis [32], pathogenic potential of these European strains, and significance in public or veterinary health remain still to be elucidated. To date, the gltA, rpoB, and ISR sequences identical with the endocarditis patient strain DNA have been identified only in the little brown myotis (Myotis lucifugus) sampled from Michigan. It seems to be possible that M. lucifugus is the only species acting as a natural reservoir for the human pathogen in North America [35]. Furthermore, B. tamiae, a newly described species isolated from patients in Thailand, has been detected in bat-associated Ixodes vespertilionis, Nycteribiidae flies, and bat spleens in Algeria [36]. However, the mode of potential transmission of bartonellae between bats and humans is still unclear.
We provide the first evidence of Rickettsia spp. in two pools of spinturnicid mites. Both identical sequences formed a unique clade comparing to other known species but can be classified as spotted fever group Rickettsia spp. Recently, Rickettsia species have been identified in several argasid tick species parasitizing bats in France, French Guiana, Mexico, Costa Rica, and the UK [11, 37,38,39,40]. However, Revees et al. [12] failed to amplify DNA of Rickettsia spp. in any of 17 pools comprising macronyssid and spinturnicid mites collected from bats mostly in North and Central America and Africa. In a Hungarian report, none of 436 mites representing the same families yielded the agent [9]. Therefore, we presume that Rickettsia spp. infections among bat-adapted mites and their hosts seem to be rather rare. The molecular presence of Rickettsia spp. in bats has been so far described only in two reports from South Africa and Brazil [41, 42]. Only one (1.7%) blood sample was positive by PCR for the msp2 gene of A. phagocytophilum, but the product could not be sequenced. On the other hand, the species-specific PCR with highly sensitive primer set allow us to claim that the positive sample could yield DNA of A. phagocytophilum. As far as we know, this is the first information indicating the possible presence of the bacterium in bats.
In conclusion, the occurrence of identical genetic variant of Bartonella among infected S. myoti samples and M. myotis bats strongly suggests that bartonellae can be shared between spinturnicid mites and their bat hosts. In this case, mites could be involved in the enzootic circulation of the bacterium as specific biological vectors, whereas greater mouse-eared bats may serve as reservoir hosts since they are capable of developing active bacteremia. On the other hand, M. myotis are unlikely to play a significant role in the maintenance of A. phagocytophilum and Rickettsia spp. Our results highlight the need for further filed studies on the vector potential of spinturnicid mites and their role in the ecology and epidemiology of bacterial infections associated with vespertilionid bats, especially regarding the genus Bartonella.
References
Billeter SA, Levy MG, Chomel BB, Breitschwerdt EB (2008) Vector transmission of Bartonella species with emphasis on the potential for tick transmission. Med Vet Entomol 22:1–15
Rudnick A (1960) A revision of the family Spinturnicidae (Acarina). Univ Calif Publ Entomol 17:157–284
Zahn A, Rupp D (2004) Ectoparasite load in European vespertilionid bats. J Zool 262:1–9
Christe P, Arlettaz R, Vogel P (2000) Variation in intensity of a parasitic mite (Spinturnix myoti) in relation to the reproductive cycle and immunocompetence of its bat host (Myotis myotis). Ecol Lett 3:207–212
Encarnação JA, Baulechner D, Becker NI (2012) Seasonal variations of wing mite infestations in male Daubenton’s bats (Myotis daubentonii) in comparison to female and juvenile bats. Acta Chiropt 14:153–159
Evans GO (1968) The external morphology of the post-embryonic developmental stages of Spinturnix myoti Kol. (Acari: Mesostigmata). Acarologia 10:589–608
Deunff J, Walter G, Bellido A, Volleth M (2004) Description of a cryptic species, Spinturnix bechsteini n. sp. (Acari, Mesostigmata, Spinturnicidaae), parasite of Myotis bechsteinii (Kuhl, 1817) (Chiroptera, Vespertilionidae) by using ecoethology of host bats and statistical methods. J Med Entomol 41:826–832
Sachanowicz K, Ciechanowski M, Piksa K (2017) Spinturnix dasycnemi (Acari: Spinturnicidae)—a poorly known Palaearctic bat mite: first records in Poland and morphometric separation from two other species of the myoti group. Ann Parasitol 63:49–56
Hornok S, Kovács R, Meli ML, Gönczi E, Hofmann-Lehmann R, Kontschán J, Gyuranecz M, Dán A, Molnár V (2012) First detection of bartonellae in a broad range of bat ectoparasites. Vet Microbiol 159:541–543
Piksa K, Stańczak J, Biernat B, Górz A, Nowak-Chmura M, Siuda K (2016) Detection of Borrelia burgdorferi sensu lato and spotted fever group rickettsiae in hard ticks (Acari, Ixodidae) parasitizing bats in Poland. Parasitol Res 115:1727–1731
Lv J, Fernández de Marco MDM, Goharriz H, Phipps LP, McElhinney LM, Hernández-Triana LM, Wu S, Lin X, Fooks AR, Johnson N (2018) Detection of tick-borne bacteria and babesia with zoonotic potential in Argas (Carios) vespertilionis (Latreille, 1802) ticks from British bats. Sci Rep 30:1865
Reeves WK, Dowling APG, Dasch GA (2006) Rickettsial agents from parasitic Dermanyssoidea (Acari: Mesostigmata). Exp Appl Acarol 38:181–188
Bogdanowicz W, Lesiński G, Sadkowska-Todys M, Gajewska M, Rutkowski R (2013) Population genetics and bat rabies: a case study of Eptesicus serotinus in Poland. Acta Chiropt 15:35–56
Frank R, Kuhn T, Werblow A, Liston A, Kochmann J, Klimpel S (2015) Parasite diversity of European Myotis species with special emphasis on Myotis myotis (Microchiroptera, Vespertilionidae) from a typical nursery roost. Parasit Vectors 8:101
Kristofik J, Danko S (2012) Arthropod ectoparasites (Acarina, Heteroptera, Diptera, Siphonaptera) of bats in Slovakia. Vespertilio 16:167–198
Arlettaz R, Ruedi M, Hausser J (1991) Field morphological identification of Myotis myotis and Myotis blythii (Chiroptera, Vespertilionidae): a multivariate approach. Myotis 29:7–16
Dusbábek F (1962) Parasitische Fledermausmilben der Tschechoslowakei I. Fam. Spinturnicidae Oudms., 1901 (Acarina, Gamasides). Acta Socent Entomol Czechoslov 59:357–380
Norman AF, Regnery R, Jameson P, Greene C, Krause DC (1995) Differentiation of Bartonella-like isolates at the species level by PCR-restriction fragment length polymorphism in the citrate synthase gene. J Clin Microbiol 33:1797–1803
Regnery RL, Spruill CL, Plikaytis BD (1991) Genotypic identification of rickettsiae and estimation of intraspecies sequence divergence for portions of two rickettsial genes. J Bacteriol 173:1576–1589
Zeidner NS, Burkot TR, Massung R, Nicholson WL, Dolan MC, Rutherford JS, Biggerstaff BJ, Maupin GO (2000) Transmission of the agent of human granulocytic ehrlichiosis by Ixodes spinipalpis ticks: evidence of an enzootic cycle of dual infection with Borrelia burgdorferi in northern Colorado. J Infect Dis 182:616–619
Massung RF, Slater KG (2003) Comparison of PCR assays for detection of the agent of human granulocytic ehrlichiosis, Anaplasma phagocytophilum. J Clin Microbiol 41:717–722
Massung RF, Slater K, Owens JH, Nicholson WL, Mather TN, Solberg VB, Olson JG (1998) Nested PCR assay for detection of granulocytic ehrlichiae. J Clin Microbiol 36:1090–1095
Chomel BB, Boulouis HJ, Maruyama S, Breitschwerdt EB (2006) Bartonella spp. in pets and effect on human health. Emerg Infect Dis 12:389–394
Postawa T, Szubert-Kruszyńska A, Ferenc H (2014) Differences between populations of Spinturnix myoti (Acari: Mesostigmata) in breeding and non-breeding colonies of Myotis myotis (Chiroptera) in central Europe: the effect of roost type. Folia Parasitol 61:581–588
Sándor AD, Földvári M, Krawczyk AI, Sprong H, Corduneanu A, Barti L, Görföl T, Estók, P, Kováts D, Szekeres S, László Z, Hornok S, Földvári G (2018) Eco-epidemiology of novel Bartonella genotypes from parasitic flies of insectivorous bats. Microb Ecol:1–13. doi:https://doi.org/10.1007/s00248-018-1195-z
Uhrin M, Kaňuch P, Krištofík J, Paule L (2010) Phenotypic plasticity in the greater mouse-eared bat in extremely different roost conditions. Acta Theriol 55:153–164
Concannon R, Wynn-Owen K, Simpson VR, Birtles RJ (2005) Molecular characterization of haemoparasites infecting bats (Microchiroptera) in Cornwall, UK. Parasitol 131:489–496
Lilley TM, Veikkolainen V, Pulliainen AT (2015) Molecular detection of Candidatus Bartonella hemsundetiensis in bats. Vector Borne Zoonotic Dis 15:706–708
Stuckey MJ, Boulouis HJ, Cliquet F, Picard-Meyer E, Servat A, Aréchiga-Ceballos N, Echevarría JE, Chomel BB (2017) Potentially zoonotic Bartonella in bats from France and Spain. Emerg Infect Dis 23:539–541
Urushadze L, Bai Y, Osikowicz LM, McKee CD, Sidamonidze K, Putkaradze D, Imnadze P, Kandaurov A, Kuzmin I, Kosoy M (2017) Prevalence, diversity, and host associations of Bartonella strains in bats from Georgia (Caucasus). PLoS Negl Trop Dis. 11:e0005428
Veikkolainen V, Vesterinen EJ, Lilley TM, Pulliainen AT (2014) Bats as reservoir hosts of human bacterial pathogen, Bartonella mayotimonensis. Emerg Infect Dis 20:960–967
Lin EY, Tsigrelis C, Baddour LM, Lepidi H, Rolain J-M, Patel R, Raoult D (2010) Candidatus Bartonella mayotimonensis and endocarditis. Emerg Infect Dis 16:500–503
Dietz C, Kiefer A (2016) Bats of Britain and Europe. Bloomsbury Natural History, London, p 400
Podsiadly E, Chmielewski T, Karbowiak G, Kedra E, Tylewska-Wierzbanowska S (2010) The occurrence of spotted fever rickettsioses and other tick-borne infections in forest workers in Poland. Vector Borne Zoonotic Dis 11:985–989
Lilley TM, Wilson CA, Bernard RF, Willcox EV, Vesterinen EJ, Webber QM, Kurpiers L, Prokkola JM, Ejotre I, Kurta A, Field KA, Reeder DM, Pulliainen AT (2017) Molecular detection of Candidatus Bartonella mayotimonensis in North American bats. Vector Borne Zoonotic Dis 17:243–246
Leulmi H, Aouadi A, Bitam I, Bessas A, Benakhla A, Raoult D, Parola P (2016) Detection of Bartonella tamiae, Coxiella burnetii and rickettsiae in arthropods and tissues from wild and domestic animals in northeastern Algeria. Parasit Vectors 9:27
Socolovschi C, Kernif T, Raoult D, Parola P (2012) Borrelia, Rickettsia, and Ehrlichia species in bat ticks, France, 2010. Emerg Infect Dis 18:1966–1975
Sánchez-Montes S, Guzmán-Cornejo C, Martínez-Nájera Y, Becker I, Venzal JM, Labruna MB (2016) Rickettsia lusitaniae associated with Ornithodoros yumatensis (Acari: Argasidae) from two caves in Yucatan, Mexico. Ticks Tick Borne Dis 7:1097–1101
Tahir D, Socolovschi C, Marié JL, Ganay G, Berenger JM, Bompar JM, Blanchet D, Cheuret M, Mediannikov O, Raoult D, Davoust B, Parola P (2016) New Rickettsia species in soft ticks Ornithodoros hasei collected from bats in French Guiana. Ticks Tick Borne Dis 7:1089–1096
Moreira-Sotoa RD, Moreira-Sotoa A, Corrales-Aguilara E, Calderón-Arguedasa O, Troyoa A (2017) Candidatus Rickettsia nicoyana: a novel Rickettsia species isolated from Ornithodoros knoxjonesi in Costa Rica. Ticks Tick Borne Dis 8:532–536
Dietrich M, Tjale MA, Weyer J, Kearney T, Seamark ECJ, Nel LH, Monadjem A, Markotter W (2016) Diversity of Bartonella and Rickettsia spp: in bats and their blood-feeding ectoparasites from South Africa and Swaziland. PLoS One 11:e0152077
Cicuttin GL, De Salvo MN, La Rosa I, Dohmen FEG (2017) Neorickettsia risticii, Rickettsia sp. and Bartonella sp. in Tadarida brasiliensis bats from Buenos Aires, Argentina. Comp Immunol Microbiol Infect Dis 52:1–5
Funding
This study was supported by the Ministry of Science and Higher Education (grant no. N N303819640).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Szubert-Kruszyńska, A., Stańczak, J., Cieniuch, S. et al. Bartonella and Rickettsia Infections in Haematophagous Spinturnix myoti Mites (Acari: Mesostigmata) and their Bat Host, Myotis myotis (Yangochiroptera: Vespertilionidae), from Poland. Microb Ecol 77, 759–768 (2019). https://doi.org/10.1007/s00248-018-1246-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-018-1246-5