Abstract
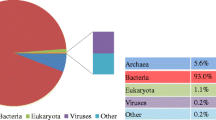
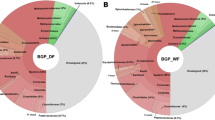
Methanogenesis from wastewater-borne organics and organic solid wastes (e.g., food residues) can be severely suppressed by the presence of toxic phenols. In this work, ambient (20 °C) and mesophilic (37 °C) methane-producing and phenol-degrading consortia were enriched and characterized using high-throughput sequencing (HTS). 454 Pyrosequencing indicated novel W22 (25.0 % of bacterial sequences) in the WWE1 and Sulfurovum-resembled species (32.0 %) in the family Campylobacterales were the most abundant in mesophilic and ambient reactors, respectively, which challenges previous knowledge that Syntrophorhabdus was the most predominant. Previous findings may underestimate bacterial diversity and low-abundance bacteria, but overestimate abundance of Syntrophorhabdus. Illumina HTS revealed that archaeal populations were doubled in ambient reactor and tripled in mesophilic reactor, respectively, compared to the ∼4.9 % (of the bacteria and archaea sequences) in the seed sludge. Moreover, unlike the dominance of Methanosarcina in seed sludge, acetotrophic Methanosaeta predominated both (71.4–76.5 % of archaeal sequences) ambient and mesophilic enrichments. Noteworthy, this study, for the first time, discovered the co-occurrence of green sulfur bacteria Chlorobia, sulfur-reducing Desulfovibrio, and Sulfurovum-resembling species under ambient condition, which could presumably establish mutualistic relationships to compete with syntrophic bacteria and methanogens, leading to the deterioration of methanogenic activity. Taken together, this HTS-based study unravels the high microbial diversity and complicated bacterial interactions within the biogas-producing and phenol-degrading bioreactors, and the identification of novel bacterial species and dominant methanogens involved in the phenol degradation provides novel insights into the operation of full-scale bioreactors for maximizing biogas generation.







Similar content being viewed by others
References
Sreekrishnan T, Kohli S, Rana V (2004) Enhancement of biogas production from solid substrates using different techniques––a review. Bioresour Technol 95:1–10
Khanal S (2009) Anaerobic biotechnology for bioenergy production: principles and applications. Wiley, North America, USA
Fang HHP, Chen T, Li YY, Chui HK (1996) Degradation of phenol in wastewater in an upflow anaerobic sludge blanket reactor. Water Res 30:1353–1360
Chen CL, Wu JH, Liu WT (2008) Identification of important microbial populations in the mesophilic and thermophilic phenol-degrading methanogenic consortia. Water Res 42:1963–1976
Fang HHP, Chan OC (1997) Toxicity of phenol towards anaerobic biogranules. Water Res 31:2229–2242
Levén L, Nyberg K, Schnürer A (2011) Conversion of phenols during anaerobic digestion of organic solid waste—a review of important microorganisms and impact of temperature. J Environ Manage. doi:10.1016/j.jenvman.2010.10.021
Veeresh GS, Kumar P, Mehrotra I (2005) Treatment of phenol and cresols in upflow anaerobic sludge blanket (UASB) process: a review. Water Res 39:154
Fang H, Liu Y, Ke S, Zhang T (2004) Anaerobic degradation of phenol in wastewater at ambient temperature. Water Sci Technol: J Int Assoc Water Pollut Res 49:95
Fang HHP, Liang DW, Zhang T, Liu Y (2006) Anaerobic treatment of phenol in wastewater under thermophilic condition. Water Res 40:427–434. doi:10.1016/j.watres.2005.11.025
Chen CL, Wu JH, Tseng IC, Liang TM, Liu WT (2009) Characterization of active microbes in a full-scale anaerobic fluidized bed reactor treating phenolic wastewater. Microbes and environments: 904220077
Boyd SA, Shelton DR, Berry D, Tiedje JM (1983) Anaerobic biodegradation of phenolic compounds in digested sludge. Appl Environ Microbiol 46:50
Zhang T, Ke S, Liu Y, Fang H (2005) Microbial characteristics of a methanogenic phenol-degrading sludge. Water Sci Technol: J Int Assoc Water Pollut Res 52:73
Levén L, Schnürer A (2010) Molecular characterisation of two anaerobic phenol-degrading enrichment cultures. Int Biodeterior Biodegrad 64:427–433. doi:10.1016/j.ibiod.2010.04.009
Shendure J, Ji H (2008) Next-generation DNA sequencing. Nat Biotechnol 26:1135–1145
Kircher M, Kelso J (2010) High-throughput DNA sequencing—concepts and limitations. BioEssays 32:524–536
Ma J, Wang Z, Yang Y, Mei X, Wu Z (2013) Correlating microbial community structure and composition with aeration intensity in submerged membrane bioreactors by 454 high-throughput pyrosequencing. Water Res 47:859–869
Lee TK, Van Doan T, Yoo K, Choi S, Kim C, Park J (2010) Discovery of commonly existing anode biofilm microbes in two different wastewater treatment MFCs using FLX titanium pyrosequencing. Appl Microbiol Biotechnol 87:2335–2343
Ishak HD, Plowes R, Sen R, Kellner K, Meyer E, Estrada DA, Dowd SE, Mueller UG (2011) Bacterial diversity in Solenopsis invicta and Solenopsis geminata ant colonies characterized by 16S amplicon 454 pyrosequencing. Microb Ecol 61:821–831
Xia Y, Cai L, Zhang T, Fang HH (2012) Effects of substrate loading and co-substrates on thermophilic anaerobic conversion of microcrystalline cellulose and microbial communities revealed using high-throughput sequencing. Int J Hydrogen Energy 37:13652–13659
Holmes DE, Risso C, Smith JA, Lovley DR (2012) Genome-scale analysis of anaerobic benzoate and phenol metabolism in the hyperthermophilic archaeon Ferroglobus placidus. The ISME J 6:146–157
EatonAD CLS, RiceEW GAE (2005) Standard Methods for the Examination of Water and Wastewater. Centennial Edition, 21st edn. American Public Health Association, Washington
Zhang T, Shao M-F, Ye L (2012) 454 Pyrosequencing reveals bacterial diversity of activated sludge from 14 sewage treatment plants. The ISME J 6:1137–1147
Qian PY, Wang Y, Lee OO, Lau SCK, Yang J, Lafi FF, Al-Suwailem A, Wong TYH (2010) Vertical stratification of microbial communities in the Red Sea revealed by 16S rDNA pyrosequencing. ISME J 5:507–518
Teske A, Sørensen KB (2007) Uncultured archaea in deep marine subsurface sediments: have we caught them all? ISME J 2:3–18
Wang Y, Zhang Y, Wang J, Meng L (2009) Effects of volatile fatty acid concentrations on methane yield and methanogenic bacteria. Biomass Bioenergy 33:848–853
Shui-zhou K, SHI Z, Tong Z, Herbert H, FANG P (2004) Degradation of phenol in an upflow anaerobic sludge blanket (UASB) reactor at ambient temperature. J Environ Sci 16
Krzywinski M, Schein J, Birol İ, Connors J, Gascoyne R, Horsman D et al (2009) Circos: an information aesthetic for comparative genomics. Genome Res 19:1639–1645
Chouari R, Le Paslier D, Dauga C, Daegelen P, Weissenbach J, Sghir A (2005) Novel major bacterial candidate division within a municipal anaerobic sludge digester. Appl Environ Microbiol 71:2145–2153
Pelletier E, Kreimeyer A, Bocs S, Rouy Z, Gyapay G, Chouari R, Rivière D, Ganesan A, Daegelen P, Sghir A (2008) “Candidatus Cloacamonas acidaminovorans”: genome sequence reconstruction provides a first glimpse of a new bacterial division. J Bacteriol 190:2572–2579
Qiu YL, Hanada S, Ohashi A, Harada H, Kamagata Y, Sekiguchi Y (2008) Syntrophorhabdus aromaticivorans gen. nov., sp. nov., the first cultured anaerobe capable of degrading phenol to acetate in obligate syntrophic associations with a hydrogenotrophic methanogen. Appl Environ Microbiol 74:2051–2058
Imachi H, Sakai S, Ohashi A, Harada H, Hanada S, Kamagata Y, Sekiguchi Y (2007) Pelotomaculum propionicicum sp. nov., an anaerobic, mesophilic, obligately syntrophic, propionate-oxidizing bacterium. Int J Syst Evol Microbiol 57:1487–1492
Balk M, Weijma J, Friedrich MW, Stams AJM (2003) Methanol utilization by a novel thermophilic homoacetogenic bacterium, Moorella mulderi sp. nov., isolated from a bioreactor. Arch Microbiol 179:315–320
Sieber JR, McInerney MJ, Gunsalus RP (2012) Genomic insights into syntrophy: the paradigm for anaerobic metabolic cooperation. Annu Rev Microbiol 66
Hernandez-Eugenio G, Fardeau ML, Patel BKC, Macarie H, Garcia JL, Ollivier B (2000) Desulfovibrio mexicanus sp. nov., a sulfate-reducing bacterium isolated from an upflow anaerobic sludge blanket (UASB) reactor treating cheese wastewaters. Anaerobe 6:305–312
Walker CB, Stolyar S, Chivian D, Pinel N, Gabster JA, Dehal PS, He Z, Yang ZK, Yen HCB, Zhou J (2009) Contribution of mobile genetic elements to Desulfovibrio vulgaris genome plasticity. Environ Microbiol 11:2244–2252
Lykidis A, Chen CL, Tringe SG, McHardy AC, Copeland A, Kyrpides NC, Hugenholtz P, Macarie H, Olmos A, Monroy O (2010) Multiple syntrophic interactions in a terephthalate-degrading methanogenic consortium. ISME J 5:122–130
Kleinsteuber S, Schleinitz KM, Breitfeld J, Harms H, Richnow HH, Vogt C (2008) Molecular characterization of bacterial communities mineralizing benzene under sulfate‐reducing conditions. FEMS Microbiol Ecol 66:143–157
Herrmann S, Kleinsteuber S, Chatzinotas A, Kuppardt S, Lueders T, Richnow HH, Vogt C (2010) Functional characterization of an anaerobic benzene‐degrading enrichment culture by DNA stable isotope probing. Environ Microbiol 12:401–411
Wüst PK, Horn MA, Drake HL (2009) Trophic links between fermenters and methanogens in a moderately acidic fen soil. Environ Microbiol 11:1395–1409
Huson DH, Auch AF, Qi J, Schuster SC (2007) MEGAN analysis of metagenomic data. Genome Res 17:377–386
Acknowledgments
The authors would like to thank GRF HKU 7190/12E for the financial support of this research. Feng Ju would like to thank the University of Hong Kong (HKU) for the postgraduate scholarship. Moreover, the authors would like to thank Dr. Feng Guo for the helpful discussion. Finally, the authors would like to thank Ms. Danping Huang for the assistance in reactor operation and Ms. Vicky Fung for the technical support.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
ESM 1
(DOCX 1225 kb)
Rights and permissions
About this article
Cite this article
Ju, F., Zhang, T. Novel Microbial Populations in Ambient and Mesophilic Biogas-Producing and Phenol-Degrading Consortia Unraveled by High-Throughput Sequencing. Microb Ecol 68, 235–246 (2014). https://doi.org/10.1007/s00248-014-0405-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-014-0405-6