Abstract
Rising sea levels and excessive water withdrawals upstream are making previously freshwater coastal ecosystems saline. Plant and animal responses to variation in the freshwater–saline interface have been well studied in the coastal zone; however, microbial community structure and functional response to seawater intrusion remains relatively unexplored. Here, we used molecular approaches to evaluate the response of the prokaryotic community to controlled changes in porewater salinity levels in freshwater sediments from the Altamaha River, Georgia, USA. This work is a companion to a previously published study describing results from an experiment using laboratory flow-through sediment core bioreactors to document biogeochemical changes as porewater salinity was increased from 0 to 10 over 35 days. As reported in Weston et al. (Biogeochemistry, 77:375–408, 62), porewater chemistry was monitored, and cores were sacrificed at 0, 9, 15, and 35 days, at which time we completed terminal restriction fragment length polymorphism and 16S rRNA clone library analyses of sediment microbial communities. The biogeochemical study documented changes in mineralization pathways in response to artificial seawater additions, with a decline in methanogenesis, a transient increase in iron reduction, and finally a dominance of sulfate reduction. Here, we report that, despite these dramatic and significant changes in microbial activity at the biogeochemical level, no significant differences were found between microbial community composition of control vs. seawater-amended treatments for either Bacterial or Archaeal members. Further, taxa in the seawater-amended treatment community did not become more “marine-like” through time. Our experiment suggests that, as seawater intrudes into freshwater sediments, observed changes in metabolic activity and carbon mineralization on the time scale of weeks are driven more by shifts in gene expression and regulation than by changes in the composition of the microbial community.







Similar content being viewed by others
References
Acinas SG, Sarma-Rupavtarm R, Klepac-Ceraj V, Polz MF (2005) PCR-induced sequence artifacts and bias: insights from comparison of two 16S rRNA clone libraries constructed from the same sample. Appl Environ Microb 71:8966–8969
Allen JA, Chambers JL, Pezeshki SR (1997) Effects of salinity on baldcypress seedlings: physiological responses and their relation to salinity tolerance. Wetlands 17:310–320
Baas-Becking LGM (1934) Geobiologie of inleiding tot de milieukunde, 18/19, Netherlands.
Baldwin DS, Rees GN, Mitchell AM, Watson G, Williams J (2006) The short-term effects of salinization on anaerobic nutrient cycling and microbial community structure in sediment from a freshwater wetland. Wetlands 26:455–464
Bernhard AE, Donn T, Giblin AE, Stahl DA (2005) Loss of diversity of ammonia-oxidizing bacteria correlates with increasing salinity in an estuary system. Environ Microbiol 7:1289–1297
Bollmann A, Laanbroek HJ (2002) Influence of oxygen partial pressure and salinity on the community composition of ammonia-oxidizing bacteria in the Schelde Estuary. Aquat Microb Ecol 28:239–247
Bouvier TC, del Giorgio PA (2002) Compositional changes in free-living bacterial communities along a salinity gradient in two temperate estuaries. Limnol Oceanogr 47:453–470
Caffrey JM, Harrington N, Solem I, Ward BB (2003) Biogeochemical processes in a small California estuary. 2. Nitrification activity, community structure and role in nitrogen budgets. Mar Ecol-Prog Ser 248:27–40
Capone DG, Kiene RP (1988) Comparison of microbial dynamics in marine and fresh-water sediments—contrasts in anaerobic carbon catabolism. Limnol Oceanogr 33:725–749
Caraco NF, Cole JJ, Likens GE (1989) Evidence for sulfate-controlled phosphorus release from sediments of aquatic systems. Nature 341:316–318
Clarke K (1993) Non-parametric multivariate analyses of changes in community structure. Aust J Ecol 18:117–143
Clarke K, Green R (1988) Statistical design and analysis for a “biological effects” study. Mar Ecol-Prog Ser 46:213–226
Cottrell MT, Kirchman DL (2004) Single-cell analysis of bacterial growth, cell size, and community structure in the Delaware Estuary. Aquat Microb Ecol 34:139–149
Crain CM, Silliman BR, Bertness SL, Bertness MD (2004) Physical and biotic drivers of plant distribution across estuarine salinity gradients. Ecology 85:2539–2549
Delong EF (1992) Archaea in coastal marine environments. Proc Natl Acad Sci U S A 89:5685–5689
Deplancke B, Hristova KR, Oakley HA, McCracken VJ, Aminov R, Mackie RI, Gaskins HR (2000) Molecular ecological analysis of the succession and diversity of sulfate-reducing bacteria in the mouse gastrointestinal tract. Appl Environ Microb 66:2166–2174
Dyszynski G, Sheldon WM (2007) RDPquery: A Java program from the Sapelo Program Microbial Observatory for automatic classification of bacterial 16S rRNA sequences based on Ribosomal Database Project taxonomy and Smith-Waterman alignment. , 2.7 ed.
Everitt B (1980) Cluster analysis, 2nd edn. Heinemann, London
Fanning JL (2003) Water use in Georgia, 2000; and trends, 1950–2000, Proceedings of the 2003 Georgia Water Resources Conference. University of Georgia, Athens
Felsenstein J (1989) PHYLIP-phylogeny inference package (version 3.2). Cladistics 5:164–166
Feseker T (2007) Numerical studies on saltwater intrusion in a coastal aquifer in northwestern Germany. Hydrogeol J 15:267–279
Francis CA, O’Mullan GD, Ward BB (2003) Diversity of ammonia monooxygenase (amoA) genes across environmental gradients in Chesapeake Bay sediments. Geobiology 1:129–140
Gardner WS, McCarthy MJ, An SM, Sobolev D, Sell KS, Brock D (2006) Nitrogen fixation and dissimilatory nitrate reduction to ammonium (DNRA) support nitrogen dynamics in Texas estuaries. Limnol Oceanogr 51:558–568
Garneau ME, Vincent WF, Alonso-Saez L, Gratton Y, Lovejoy C (2006) Prokaryotic community structure and heterotrophic production in a river-influenced coastal arctic ecosystem. Aquat Microb Ecol 42:27–40
Gillanders BM, Kingsford MJ (2002) Impact of changes in flow of freshwater on estuarine and open coastal habitats and the associated organisms. Oceanogr Mar Biol 40:233–309
Gilmore RG (1995) Environmental and biogeographic factors influencing ichthyofaunal diversity—Indian-River Lagoon. B Mar Sci 57:153–170
Giovannoni SJ (1991) The polymerase chain reaction. In: Stackebrandt E, Goodfellow M (eds) Nucleic acid techniques in bacterial systematics. Wiley, New York
Gleick PH (2003) Water use. Annu Rev Env Resour 28:275–314
Hales BA, Edwards C, Ritchie DA, Hall G, Pickup RW, Saunders JR (1996) Isolation and identification of methanogen-specific DNA from blanket bog feat by PCR amplification and sequence analysis. Appl Environ Microb 62:668–675
Hall T (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Henriksen JR (2004) webLIBSHUFF, http://libshuff.mib.uga.edu.
Henriques IS, Almeida A, Cunha A, Correia A (2004) Molecular sequence analysis of prokaryotic diversity in the middle and outer sections of the Portuguese estuary Ria de Aveiro. FEMS Microbiol Ecol 49:269–279
Hewson I, Vargo GA, Fuhrman JA (2003) Bacterial diversity in shallow oligotrophic marine benthos and overlying waters: effects of virus infection, containment, and nutrient enrichment. Microbial Ecol 46:322–336
Jackson RB, Carpenter SR, Dahm CN, McKnight DM, Naiman RJ, Postel SL, Running SW (2001) Water in a changing world. Ecol Appl 11:1027–1045
James KR, Cant B, Ryan T (2003) Responses of freshwater biota to rising salinity levels and implications for saline water management: a review. Aust J Bot 51:703–713
Jukes TH, Cantor CR (1969) Evolution of protein molecules, p. 21–132. In: Munro HN (ed) Mammalian protein metabolism. Academic, New York
Kaufmann GF, Sartorio R, Lee SH, Rogers CJ, Meijler MM, Moss JA, Clapham B, Brogan AP, Dickerson TJ, Janda KD (2005) Revisiting quorum sensing: discovery of additional chemical and biological functions for 3-oxo-N-acylhomoserine lactones. Proc Natl Acad Sci U S A 102:309–314
Knowles N, Cayan DR (2004) Elevational dependence of projected hydrologic changes in the San Francisco Estuary and watershed. Clim Change 62:319–336
Kreth J, Merritt J, Shi WY, Qi FX (2005) Competition and coexistence between Streptococcus mutans and Streptococcus sanguinis in the dental biofilm. J Bacteriol 187:7193–7203
Langenheder S, Kisand V, Wikner J, Tranvik LJ (2003) Salinity as a structuring factor for the composition and performance of bacterioplankton degrading riverine DOC. FEMS Microbiol Ecol 45:189–202
Liu WT, Marsh TL, Cheng H, Forney LJ (1997) Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl Environ Microb 63:4516–4522
Magurran A (1988) Ecological diversity and its measurement. Princeton University Press, Princeton
Martiny JBH, Bohannan BJM, Brown JH, Colwell RK, Fuhrman JA, Green JL, Horner-Devine MC, Kane M, Krumins JA, Kuske CR, Morin PJ, Naeem S, Ovreas L, Reysenbach AL, Smith VH, Staley JT (2006) Microbial biogeography: putting microorganisms on the map. Nat Rev Microbiol 4:102–112
Meyer JL, Sale MJ, Mulholland PJ, Poff NL (1999) Impacts of climate change on aquatic ecosystem functioning and health. J Am Water Resour As 35:1373–1386
Mishra SR, Pattnaik P, Sethunathan N, Adhya TK (2003) Anion-mediated salinity affecting methane production in a flooded alluvial soil. Geomicrobiol J 20:579–586
Mulholland PJ, Best GR, Coutant CC, Hornberger GM, Meyer JL, Robinson PJ, Stenberg JR, Turner RE, VeraHerrera F, Wetzel RG (1997) Effects of climate change on freshwater ecosystems of the south-eastern United States and the Gulf Coast of Mexico. Hydrol Process 11:949–970
Muthumbi W, Boon N, Boterdaele R, De Vreese I, Top EM, Verstraete W (2001) Microbial sulfate reduction with acetate: process performance and composition of the bacterial communities in the reactor at different salinity levels. Appl Microbiol Biot 55:787–793
Pattnaik P, Mishra SR, Bharati K, Mohanty SR, Sethunathan N, Adhya TK (2000) Influence of salinity on methanogenesis and associated microflora in tropical rice soils. Microbiol Res 155:215–220
Pett-Ridge J, Silver WL, Firestone MK (2006) Redox fluctuations frame microbial community impacts on N-cycling rates in a humid tropical forest soil. Biogeochemistry 81:95–110
Piccini C, Conde D, Alonso C, Sommaruga R, Pernthaler J (2006) Blooms of single bacterial species in a coastal lagoon of the southwestern Atlantic Ocean. Appl Environ Microb 72:6560–6568
Ranjan SP, Kazama S, Sawamoto M (2006) Effects of climate and land use changes on groundwater resources in coastal aquifers. J Environ Manage 80:25–35
Rysgaard S, Thastum P, Dalsgaard T, Christensen PB, Sloth NP (1999) Effects of salinity on NH4+ adsorption capacity, nitrification, and denitrification in Danish estuarine sediments. Estuaries 22:21–30
Santoro AE, Boehm AB, Francis CA (2006) Denitrifier community composition along a nitrate and salinity gradient in a coastal aquifer. Appl Environ Microb 72:2102–2109
Schloss PD, Handelsman J (2005) Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl Environ Microb 71:1501–1506
Selje N, Simon M (2003) Composition and dynamics of particle-associated and free-living bacterial communities in the Weser estuary, Germany. Aquat Microb Ecol 30:221–237
Short SM, Jenkins BD, Zehr JP (2004) Spatial and temporal distribution of two diazotrophic bacteria in the Chesapeake Bay. Appl Environ Microb 70:2186–2192
Singleton DR, Furlong MA, Rathbun SL, Whitman WB (2001) Quantitative comparisons of 16S rRNA gene sequence libraries from environmental samples. Appl Environ Microb 67:4374–4376
Smith JM, Green SJ, Kelley CA, Prufert-Bebout L, Bebout BM (2008) Shifts in methanogen community structure and function associated with long-term manipulation of sulfate and salinity in a hypersaline microbial mat. Environ Microbiol 10:386–394
Stepanauskas R, Moran MA, Bergamaschi BA, Hollibaugh JT (2003) Covariance of bacterioplankton composition and environmental variables in a temperate delta system. Aquat Microb Ecol 31:85–98
Thompson JD, Higgins DG, Gibson TJ (1994) Clustal-W—improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Tobias CR, Anderson IC, Canuel EA, Macko SA (2001) Nitrogen cycling through a fringing marsh-aquifer ecotone. Mar Ecol-Prog Ser 210:25–39
Weston NB, Dixon RE, Joye SB (2006) Ramifications of increased salinity in tidal freshwater sediments: Geochemistry and microbial pathways of organic matter mineralization. J Geophys Res 111
Weston NB, Hollibaugh JT, Joye SB (2009) Population growth away from the coastal zone: Thirty years of land use change and nutrient export in the Altamaha River, GA. Sci Total Environ 407:3347–3356
Weston NB, Porubsky WP, Samarkin VA, Erickson M, Macavoy SE, Joye SB (2006) Porewater stoichiometry of terminal metabolic products, sulfate, and dissolved organic carbon and nitrogen in estuarine intertidal creek-bank sediments. Biogeochemistry 77:375–408
Zimmerman AR, Benner R (1994) Denitrification, nutrient regeneration and carbon mineralization in sediments of Galveston Bay, Texas, USA. Mar Ecol-Prog Ser 114:275–288
Acknowledgments
This work was funded by a NSF Microbial Biology post-doctoral fellowship (to JWE), by a grant from the Gordon and Betty Moore Foundation (to MAM), and by the National Science Foundations’s Georgia Coastal Ecosystems Long Term Ecological Research Program (OCE 99-82133 to SBJ). We thank J. Menken and R. Dixon for laboratory assistance. Molecular analysis support was graciously provided by T. Pickering and W. Ye.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
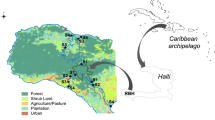
Supplemental Material Figure 1
Study site map indicating sampling location with respect to the coastal Altamaha River complex and the eastern North American coast (inset). (DOC 229 kb)
Supplemental Material Figure 2
Bootstrapped, neighbor-joining tree using all nonredundant sequences collected from the experiment. Our sequences were aligned with sequences from the SIMO database (name includes “SIMO”) as well as sequences of known Bacterial species (named in tree). Sequences collected from the initially sacrificed core are labeled “Initial”. Sequences from the salinity-amended samples have “RT” in the name, versus the unamended samples, which have “CT” in the name. Samples taken on day 9 are labeled “15”, samples from day 15 are labeled “23”, and samples taken from day 35 are labeled “41”. (PDF 3.11 MB)
Rights and permissions
About this article
Cite this article
Edmonds, J.W., Weston, N.B., Joye, S.B. et al. Microbial Community Response to Seawater Amendment in Low-Salinity Tidal Sediments. Microb Ecol 58, 558–568 (2009). https://doi.org/10.1007/s00248-009-9556-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-009-9556-2