Abstract
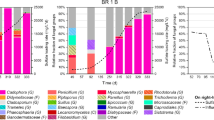
Waste streams from industrial processes such as metal smelting or mining contain high concentrations of sulfate and metals with low pH. Dissimilatory sulfate reduction carried out by sulfate-reducing bacteria (SRB) at low pH can combine sulfate reduction with metal-sulfide precipitation and thus open possibilities for selective metal recovery. This study investigates the microbial diversity and population changes of a single-stage sulfidogenic gas-lift bioreactor treating synthetic zinc-rich waste water at pH 5.5 by denaturing gradient gel electrophoresis of 16S rRNA gene fragments and quantitative polymerase chain reaction. The results indicate the presence of a diverse range of phylogenetic groups with the predominant microbial populations belonging to the Desulfovibrionaceae from δ-Proteobacteria. Desulfovibrio desulfuricans-like populations were the most abundant among the SRB during the three stable phases of varying sulfide and zinc concentrations and increased from 13% to 54% of the total bacterial populations over time. The second largest group was Desulfovibrio marrakechensis-like SRB that increased from 1% to about 10% with decreasing sulfide concentrations. Desulfovibrio aminophilus-like populations were the only SRB to decrease in numbers with decreasing sulfide concentrations. However, their population was <1% of the total bacterial population in the reactor at all analyzed time points. The number of dissimilatory sulfate reductase (DsrA) gene copies per number of SRB cells decreased from 3.5 to 2 DsrA copies when the sulfide concentration was reduced, suggesting that the cells' sulfate-reducing capacity was also lowered. This study has identified the species present in a single-stage sulfidogenic bioreactor treating zinc-rich wastewater at low pH and provides insights into the microbial ecology of this biotechnological process.



Similar content being viewed by others
References
Amann RI, Ludwig W, Schleifer KH (1995) Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev 59:143–169
Baena S, Fardeau ML, Labat M, Ollivier B, Garcia JL, Patel BK (1998) Desulfovibrio aminophilus sp. nov., a novel amino acid degrading and sulfate reducing bacterium from an anaerobic dairy wastewater lagoon. Syst Appl Microbiol 21:498–504
Bijmans, MFM, Ennin, F, Dopson, M, Lens, PNL, Buisman, CJN (2009) Effect of sulfide removal on sulfate reduction at pH 5 in a hydrogen fed gas-lift bioreactor. J Microbiol Biotechnol 18:1809–1818
Bijmans MFM, Peeters TWT, Lens PNL, Buisman CJN (2008) High rate sulfate reduction at pH 6 in a pH-auxostat submerged membrane bioreactor fed with formate. Water Res 42:2439–2448
Bijmans MFM, Van Helvoort PJ, Buisman CJN, Lens P (2009) Effect of the sulfide concentration on zinc bio-precipitation in a single stage sulfidogenic bioreactor at pH 5.5 (submitted)
Brysch K, Schneider C, Fuchs G, Widdel F (1987) Lithoautotrophic growth of sulfate-reducing bacteria, and description of Desulfobacterium autotrophicum gen. nov., sp. nov. Arch Microbiol 148:264–274
Chamkh, F, El Amrani, K, Lemos, PC, Besson, S, Lorquin, J, Fassouane, A, Reis, M, Qatibi, AI (2007) Desulfovibrio marrakechensis sp. nov., a new sulfate-reducing bacterium isolated from olive mill wastewater evaporation ponds. http://www.ncbi.nlm.nih.gov Unpublished
Chang YJ, Peacock AD, Long PE, Stephen JR, McKinley JP, Macnaughton SJ, Hussain AK, Saxton AM, White DC (2001) Diversity and characterization of sulfate-reducing bacteria in groundwater at a uranium mill tailings site. Appl Environ Microbiol 67:3149–3160
Colleran E, Finnegan S, Lens P (1995) Anaerobic treatment of sulphate-containing waste streams. Antonie van Leeuwenhoek 67:29–46
Dar SA, Kuenen JG, Muyzer G (2005) Nested PCR-denaturing gradient gel electrophoresis approach to determine the diversity of sulfate-reducing bacteria in complex microbial communities. Appl Environ Microbiol 71:2325–2330
Dojka MA, Hugenholtz P, Haack SK, Pace NR (1998) Microbial diversity in a hydrocarbon- and chlorinated-solvent-contaminated aquifer undergoing intrinsic bioremediation. Appl Environ Microbiol 64:3869–3877
Dopson M, Lindstrom EB (2004) Analysis of community composition during moderately thermophilic bioleaching of pyrite, arsenical pyrite, and chalcopyrite. Microbial Ecol 48:19–28
Gibson GR (1990) Physiology and ecology of the sulphate-reducing bacteria. J Appl Bacteriol 69:769–797
Isa Z, Grusenmeyer S, Verstraete W (1986) Sulfate reduction relative to methane production in high-rate anaerobic digestion: microbiological aspects. Appl Environ Microbiol 51:580–587
Johnson DB, Hallberg KB (2005) Acid mine drainage remediation options: a review. Sci Tot Environ 338:3–14
Jong T, Parry DL (2006) Microbial sulfate reduction under sequentially acidic conditions in an upflow anaerobic packed bed bioreactor. Water Res 40:2561–2571
Kaksonen AH, Plumb JJ, Franzmann PD, Puhakka JA (2004) Simple organic electron donors support diverse sulfate-reducing communities in fluidized-bed reactors treating acidic metal- and sulfate-containing wastewater. FEMS Microbiol Ecol 47:279–289
Kaksonen AH, Plumb JJ, Robertson WJ, Franzmann PD, Gibson JAE, Puhakka JA (2004) Culturable diversity and community fatty acid profiling of sulfate-reducing fluidized-bed reactor treating acidic, metal-containing wastewater. Geomicrobiology J 21:469–480
Kondo R, Nedwell DB, Purdy KJ, de Queiroz Silva S (2004) Detection and enumeration of sulfate-reducing bacteria in estuarine marine sediments by competitive PCR. Geomicrobiol J 21:145–157
Koschorreck M, Wendt-Potthoff K, Geller W (2003) Microbial sulfate reduction at low pH in sediments of an acidic lake in Argentina. Environ Sci Technol 37:1159–1162
Kusel K, Dorsch T, Acker G, Stackebrandt E, Drake H (2000) Clostridium scatologenes strain SL1 isolated as an acetogenic bacterium from acidic sediments. Int J System Evol Microbiol 50:537–546
Lens PNL, Visser A, Janssen AJH, Hulshoff Pol LL, Lettinga G (1998) Biotechnological treatment of sulfate-rich wastewaters. Crit Rev Environ Sci Technol 28:41–88
Liou JSC, Balkwill DL, Drake GR, Tanner RS (2005) Clostridium carboxidivorans sp. nov., a solvent-producing clostridium isolated from an agricultural settling lagoon, and reclassification of the acetogen Clostridium scatologenes strain SL1 as Clostridium drakei sp. nov. Int J System Evol Microbiol 55:2085–2091
Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar Buchner A, Lai T, Steppi S, Jobb G, Forster W, Brettske I, Gerber S, Ginhart AW, Gross O, Grumann S, Hermann S, Jost R, Konig A, Liss T, Lussmann R, May M, Nonhoff B, Reichel B, Strehlow R, Stamatakis A, Stuckmann N, Vilbig A, Lenke M, Ludwig T, Bode A, Schleifer KH (2004) ARB: a software environment for sequence data. Nucleic Acids Res 32:1363–1371
Maillacheruvu KY, Parkin GF (1996) Kinetics of growth, substrate utilization and sulfide toxicity for propionate, acetate, and hydrogen utilizers in anaerobic systems. Water Environ Res 68:1099–1106
Manz W, Wagner M, Amann R, Schleifer KH (1994) In situ characterization of the microbial consortia active in two wastewater treatment plants. Water Res 28:1715–1723
McGinnis S, Madden TL (2004) BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res 32:W20–W25
Monis PT, Giglio S, Saint CP (2005) Comparison of SYTO9 and SYBR Green I for real-time polymerase chain reaction and investigation of the effect of dye concentration on amplification and DNA melting curve analysis. Anal Biochem 340:24–34
Morales TA, Dopson M, Athar R, Herbert RB (2005) Analysis of bacterial diversity in acidic pond water and compost after treatment of artificial acid mine drainage for metal removal. Biotechnol Bioengin 90:543–551
Muyzer G, de Waal EC, Uitterlinden AG (1993) Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 59:695–700
Muyzer G, Teske A, Wirsen CO, Jannasch HW (1995) Phylogenetic relationships of Thiomicrospira species and their identification in deep-sea hydrothermal vent samples by denaturing gradient gel electrophoresis of 16S rDNA fragments. Arch Microbiol 164:165–172
Nakagawa T, Hanada S, Maruyama A, Marumo K, Urabe T, Fukui M (2002) Distribution and diversity of thermophilic sulfate-reducing bacteria within a Cu-Pb-Zn mine (Toyoho, Japan). FEMS Microbiol Ecol 41:199–209
O'Flaherty V, Mahony T, O'Kennedy RE, Colleran E (1998) Effect of pH on growth kinetics and sulfide toxicity thresholds of a range of methanogenic, syntrophic and sulfate-reducing bacteria. Process Biochem 33:555–569
Okabe S, Nielsen PH, Jones WL, Characklis WG (1995) Sulfide product inhibition of Desulfovibrio desulfuricans in batch and continuous cultures. Water Res 29:571–578
Oude Elferink SJWH, Visser A, Pol LWH, Stams AJM (1994) Sulfate reduction in methanogenic bioreactors. FEMS Microbiol Rev 15:119–136
Pol LW, Lens PN, Weijma J, Stams AJ (2001) New developments in reactor and process technology for sulfate reduction. Water Sci Technol 44:67–76
Sassi AB, Boularbah A, Jaouad A, Walker G, Boussaid A (2006) A comparison of olive oil mill wastewaters (OMW) from three different processes in morocco. Process Biochem 41:74–78
Schäfer H, Muyzer G (2001) Denaturing gradient gel electrophoresis in marine microbial ecology. Methods in microbiology. In: Paul JH (ed) Marine microbiology, vol 30. Academic, New York, pp 425–468
Stams AJM, Plugge CM, de Bok FAM, van Houten BHGW, Lens P, Dijkman H, Weijma J (2005) Metabolic interactions in methanogenic and sulfate reducing bacteria. Water Sci Technol 52:13–20
von Wintzingerode F, Gobel UB, Stackebrandt E (1997) Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol Rev 21:213–229
Weijma J, Copini CFM, Buisman CJN, Schultz CE (2002) Biological recovery of metals, sulfur and water in the mining and metallurgical industry. In: Lens PNL, Hulshoff Pol L (eds) Water recycling and resource recovery in industry: analysis, technologies and implementation. IWA, London, pp 605–625
Weijma J, Gubbels F, Hulshoff Pol LW, Stams AJM, Lens P, Lettinga G (2002) Competition for H2 between sulfate reducers, methanogens and homoacetogens in a gas-lift reactor. Water Sci Technol 45:75–80
Widdel F (1988) Microbiology and ecology of sulfate-and sulfur-reducing bacteria, p. 469–585. In: Zehnder AJB (ed) Biology of anaerobic microorganisms. Wiley, New York
Widdel F, Bak F (1992) Gram-negative mesophilic sulfate-reducing bacteria. In: Balows A, Trüper HG, Dworkin M, Harder W, Schleifer K-H (eds) The prokaryotes, 2nd edn. Springer, New York, pp 3352–3378
Zhang T, Fang HH (2006) Applications of real-time polymerase chain reaction for quantification of microorganisms in environmental samples. Appl Microbiol Biotechnol 70:281–289
Acknowledgments
This work was carried out in the frame of European Sixth Framework Programme for Research and Development “BioMinE” project (European contract NMP1-CT-500329-1).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Dar, S.A., Bijmans, M.F.M., Dinkla, I.J.T. et al. Population Dynamics of a Single-Stage Sulfidogenic Bioreactor Treating Synthetic Zinc-Containing Waste Streams. Microb Ecol 58, 529–537 (2009). https://doi.org/10.1007/s00248-009-9509-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-009-9509-9