Abstract
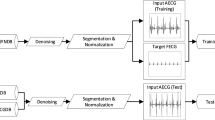
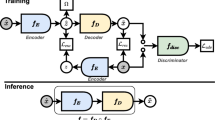
To extract weak fetal ECG signals from the mixed ECG signal on the mother's abdominal wall, providing a basis for accurately estimating fetal heart rate and analyzing fetal ECG morphology. First, based on the relationship between the maternal chest ECG signal and the maternal ECG component in the abdominal signal, the temporal convolutional encoder-decoder network (TCED-Net) model is trained to fit the nonlinear transmission of the maternal ECG signal from the chest to the abdominal wall. Then, the maternal chest ECG signal is nonlinearly transformed to estimate the maternal ECG component in the abdominal mixed signal. Finally, the estimated maternal ECG component is subtracted from the abdominal mixed signal to obtain the fetal ECG component. The simulation results on the FECGSYN dataset show that the proposed approach achieves the best performance in F1 score, mean square error (MSE), and quality signal-to-noise ratio (qSNR) (98.94%, 0.18, and 8.30, respectively). On the NI-FECG dataset, although the fetal ECG component is small in energy in the mixed signal, this method can effectively suppress the maternal ECG component and thus extract a clearer and more accurate fetal ECG signal. Compared with existing algorithms, the proposed method can extract clearer fetal ECG signals, which has significant application value for effective fetal health monitoring during pregnancy.






Similar content being viewed by others
Availability of Data and Materials
All data, models, and code generated or used during the study appear in the submitted article.
References
Fotiadou E, Sloun R, Laar J et al (2021) A dilated inception CNN-LSTM network for fetal heart rate estimation. Physiol Meas 42(4):045007
Clifford GD, Silva I, Behar J et al (2014) Non-invasive fetal ECG analysis. Physiol Meas 35(8):1521
Shaw CJ, Lees CC, Giussani DA (2016) Variations on fetal heart rate variability. J Physiol 594(5):1279
Andreotti F, Grer F, Malberg H et al (2017) Non-Invasive Fetal ECG Signal Quality Assessment for Multichannel Heart Rate Estimation. IEEE Trans Biomed Eng 12:1–1
Zhou Z, Huang K, Qiu Y et al (2021) Morphology extraction of fetal electrocardiogram by slow-fast LSTM network. Biomed Signal Process Control 68:102664
Kahankova R, Martinek R, Jaros R et al (2019) A review of signal processing techniques for non-invasive fetal electrocardiography. IEEE Rev Biomed Eng 13:51–73
Widrow B, Glover JR, McCool JM, Kaunitz J, Williams CS, Hearn RH, Zeidler JR, Dong JE, Goodlin RC (1975) Adaptive noise cancelling: Principles and applications. Proc IEEE 63(12):1692–1716
Kanjilal PP, Palit S, Saha G (1997) Fetal ECG extraction from single-channel maternal ECG using singular value decomposition. IEEE Trans Biomed Eng 44(1):51–59
Bergveld P, Meijer WJ (1981) A new technique for the suppression of the MECG. IEEE Trans Biomed Eng 4:348–354
Martens SM, Rabotti C, Mischi M, Sluijter RJ (2007) A robust fetal ECG detection method for abdominal recordings. Physiol Meas 28(4):373
Behar J, Andreotti F, Oster J, Clifford GD (2014) A Bayesian filtering framework for accurate extracting of the non-invasive FECG morphology. In: Computing in cardiology 2014, 2014. IEEE. pp. 53–56
Matonia A., Jezewski J., Horoba K., Gacek A., Labaj P (2006) The maternal ECG suppression algorithm for efficient extraction of the fetal ECG from abdominal signal. In: 2006 International Conference of the IEEE Engineering in Medicine and Biology Society. IEEE, pp 3106–3109
Niknazar M, Rivet B, Jutten C (2013) Fetal ECG extraction by extended state Kalman filtering based on single-channel recordings. IEEE Trans Biomed Eng 60(5):1345–1352
Sarafan S et al (2022) A Novel ECG Denoising Scheme Using the Ensemble Kalman Filter. In 2022 44th Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC). IEEE, pp 2005–2008
Sutha P, Jayanthi VE (2018) Fetal electrocardiogram extraction and analysis using adaptive noise cancellation and wavelet transformation techniques. J Med Syst 42:1–18
Mohammed Kaleem A, Kokate RD (2021) A survey on FECG extraction using neural network and adaptive filter. Soft Comput 25(6):4379–4392
Xue J, Yu L (2021) Applications of machine learning in ambulatory ECG. Hearts 2(4):472–494
Zhong W, Liao L, Guo X et al (2019) Fetal electrocardiography extraction with residual convolutional encoder–decoder networks. Australas Phys Eng Sci Med 42(4):1081–1089
Wei Xu, Wei Z, Wei Q et al (2020) Research and application of a deep learning model. Comput Technol Dev 30(7):5
Rasti-Meymandi A, Ghaffari A (2021) AECG-DecompNet: abdominal ECG signal decomposition through deep-learning model. Physiol Meas 42(4):045002
Behar J, Andreotti F, Zaunseder S et al (2014) An ECG simulator for generating maternal-foetal activity mixtures on abdominal ECG recordings. Physiol Meas 35(8):1537
PhysioBank PT (2000) PhysioNet: components of a new research resource for complex physiologic signals. Circulation 101(23):215–220
Widrow B, Mccool JM Jr et al (1976) Adaptive Noise Cancelling: Principles and Applications. Proceedings of the IEEE 63(12):1692–1716
Ifeachor EC et al (2004) Nonlinear methods for biopattern analysis: role and challenges. In: The 26th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, IEEE, pp 5400–5406
Ma Y, Xiao Y, Wei G et al (2018) Foetal ECG extraction using non-linear adaptive noise canceller with multiple primary channels. IET Signal Proc 12(2):219–227
Qiu Y (2019) Research on fetal ECG extraction method based on recurrent neural network. Zhejiang University, Zhejiang
Wang X, Xiaoyang Z, Xinying W et al (2022) Super resolution reconstruction of single image based on non decimated wavelet edge learning depth residual network. J Electron 50(7):1753–1765
Zhang F, Cai N, Wu J et al (2018) Image denoising method based on a deep convolution neural network. IET Image Proc 12(4):485–493
Hewage P, Trovati M, Pereira E et al (2021) Deep learning-based effective fine-grained weather forecasting model. Pattern Anal Appl 24(1):343–366
Yuqing Y, Jianghui C, Haifeng Y et al (2022) LAMOST low-quality spectral analysis based on influence space and data field. Spectrosc Spectral Anal 42(4):1186–1191
Billeci L, Varanini M (2017) A combined independent source separation and quality index optimization method for fetal ECG extraction from abdominal maternal leads. Sensors 17(5):1135
Behar J, Johnson A, Clifford GD et al (2014) A comparison of single channel fetal ECG extraction methods. Ann Biomed Eng 42(6):1340–1353
Acknowledgements
We would like to express our gratitude to Dr. Yisheng Wu and Dr. Shangping Chen for their valuable insights and suggestions during the development of this research project. We also thank the staff at Zhaoqing First People's Hospital for their assistance with data collection and analysis.
Funding
This work was supported in part by the Medical Science and Technology Research Fund of Guangdong Province (B2020199, B2023392), Guangdong Province Continuing Education Quality Improvement Project (JXJYGC2022GX543), Characteristic Innovation Projects of Ordinary University in Guangdong Province (2019GKTSCX129), Guangdong Vocational College Teaching Management Steering Committee Education and Teaching Reform Key Project (YJXGLW2022Z10), Guangdong Higher Education Association “14th Five-Year” Plan 2023 Higher Education Research Key Project (23GZD22), Scientific research planning project of Guangdong Vocational and Technical Education Association (202212G247), and The Medical Simulation Education Teaching Research Project of Guangdong Provincial Higher Vocational Education Medical and Health Professional Teaching Steering Committee (2022MYLX081).
Author information
Authors and Affiliations
Contributions
HH completed this study on her own.
Corresponding author
Ethics declarations
Competing interests
None.
Ethical Approval
This study involving human participants, human material, or human data, was performed in accordance with the Declaration of Helsinki. The experimental protocols were approved by the Institutional Review Board (IRB) of Zhaoqing Medical College (Approval No. 2022-0124).
Consent to Participate
Informed consent was obtained from all participants prior to their inclusion in the study.
Consent for Publication
Informed consent for the publication of identifying information and images,which could lead to the identification of study participants, was obtained from all subjects. All participants agreed to the publication of their anonymized data in this online open-access publication.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Huang, H. A Novel Approach to Fetal ECG Extraction Using Temporal Convolutional Encoder–Decoder Network (TCED-Net). Pediatr Cardiol 44, 1726–1735 (2023). https://doi.org/10.1007/s00246-023-03273-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00246-023-03273-z