Abstract
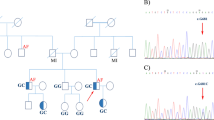
Idiopathic ventricular tachycardia (IVT) is the major cause of sudden cardiac death. Patients with IVT were usually manifested without structural heart disease. In this present study, we performed family-based whole genome sequencing (WGS) and Sanger sequencing for a 5-year-old Chinese boy with IVT and all the unaffected family members in order to identify the candidate gene and disease-causing mutation underlying the disease phenotype. Results showed that a novel heterozygous single-nucleotide duplication (c.128dup) and a novel heterozygous missense (c.3328A > G) variant in ABCA5 gene were identified in the proband. The single-nucleotide duplication (c.128dupT), inherited from his father and patrilineal grandfather, leads to a frameshift which results into the formation of a truncated ABCA5 protein of 50 (p.Leu43Phefs*8) amino acids. Hence, it is a loss-of-function mutation. The missense (c.3328A > G) variant, inherited from his mother, leads to the replacement of isoleucine by valine at the position of 1110 (p.Ile1110Val) of the ABCA5 protein. Multiple sequence alignment showed that p.Ile1110 is evolutionarily conserved among several species indicating both the structural and functional significance of the p.Ile1110 residue in the wild-type ABCA5 protein. Quantitative RT-PCR showed that the ABCA5 mRNA expression levels were decreased in the proband. These two novel variants of ABCA5 gene were co-segregated well among all the members of this family. Our present study also strongly supports the importance of using family-based whole genome sequencing for identifying novel candidate genes associated with IVT.





Similar content being viewed by others
Abbreviations
- IVT:
-
Idiopathic ventricular tachycardia
- WGS:
-
Whole genome sequencing
- WES:
-
Whole exome sequencing
- SNV:
-
Single-nucleotide variant
- INDEL:
-
Insertion/deletion
- CHD:
-
Congenital heart disease
- SCD:
-
Sudden cardiac death
- VF:
-
Ventricular fibrillation
- ECG:
-
Electrocardiographic
- ACMG:
-
American college of medical genetics and genomics
- VUS:
-
Variant of uncertain significance
- DCM:
-
Dilated cardiomyopathy
References
Glessner JT, Bick AG, Ito K, Homsy JG, Rodriguez-Murillo L, Fromer M, Mazaika E, Vardarajan B, Italia M, Leipzig J (2014) Increased frequency of de novo copy number variants in congenital heart disease by integrative analysis of single nucleotide polymorphism array and exome sequence data. Circ Res 115:884–896
Fahed AC, Gelb BD, Seidman J, Seidman CE (2013) Genetics of congenital heart disease: the glass half empty. Circ Res 112:707–720
van der Linde D, Konings EE, Slager MA, Witsenburg M, Helbing WA, Takkenberg JJ, Roos-Hesselink JW (2011) Birth prevalence of congenital heart disease worldwide: a systematic review and meta-analysis. J Am Coll Cardiol 58:2241–2247
Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, Aboyans V, Abraham J, Adair T, Aggarwal R, Ahn SY (2012) Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the global burden of disease study 2010. The lancet 380:2095–2128
Pierpont ME, Basson CT, Benson DW Jr, Gelb BD, Giglia TM, Goldmuntz E, McGee G, Sable CA, Srivastava D, Webb CL (2007) Genetic basis for congenital heart defects: current knowledge: a scientific statement from the American heart association congenital cardiac defects committee, council on cardiovascular disease in the young: endorsed by the American academy of pediatrics. Circulation 115:3015–3038
Ng SB, Bigham AW, Buckingham KJ, Hannibal MC, McMillin MJ, Gildersleeve HI, Beck AE, Tabor HK, Cooper GM, Mefford HC (2010) Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat Genet 42:790
Homsy J, Zaidi S, Shen Y, Ware JS, Samocha KE, Karczewski KJ, DePalma SR, McKean D, Wakimoto H, Gorham J (2015) De novo mutations in congenital heart disease with neurodevelopmental and other congenital anomalies. Science 350:1262–1266
Zaidi S, Choi M, Wakimoto H, Ma L, Jiang J, Overton JD, Romano-Adesman A, Bjornson RD, Breitbart RE, Brown KK (2013) De novo mutations in histone-modifying genes in congenital heart disease. Nature 498:220
Zipes DP, Camm AJ, Borggrefe M, Buxton AE, Chaitman B, Fromer M, Gregoratos G, Klein G, Myerburg RJ, Quinones MA (2006) ACC/AHA/ESC 2006 guidelines for management of patients with ventricular arrhythmias and the prevention of sudden cardiac death: a report of the American college of cardiology/American heart association task force and the European society of cardiology committee for practice guidelines (writing committee to develop guidelines for management of patients with ventricular arrhythmias and the prevention of sudden cardiac death). J Am Coll Cardiol 48:e247–e346
Vereckei A, Duray G, Szénási G, Altemose GT, Miller JM (2008) New algorithm using only lead aVR for differential diagnosis of wide QRS complex tachycardia. Heart rhythm 5:89–98
Crawford T, Mueller G, Good E, Jongnarangsin K, Chugh A, Pelosi F Jr, Ebinger M, Oral H, Morady F, Bogun F (2010) Ventricular arrhythmias originating from papillary muscles in the right ventricle. Heart rhythm 7:725–730
Lerman BB (2007) Mechanism of outflow tract tachycardia. Heart Rhythm 4:973–976
Kim RJ, Iwai S, Markowitz SM, Shah BK, Stein KM, Lerman BB (2007) Clinical and electrophysiological spectrum of idiopathic ventricular outflow tract arrhythmias. J Am Coll Cardiol 49:2035–2043
Yan A, Shayne A, Brown K, Gupta S, Chan C, Luu T, Di Carli M, Reynolds H, Stevenson W, Kwong R (2006) Characterization of the Peri-Infarct Zone by contrast-enhanced cardiac MRI is a powerful predictor of post-myocardial infarction mortality. Circulation 114:32–39
Gaita F, Giustetto C, Di Donna P, Richiardi E, Libero L, Brusin MCR, Molinari G, Trevi G (2001) Long-term follow-up of right ventricular monomorphic extrasystoles. J Am Coll Cardiol 38:364–370
Dewey FE, Grove ME, Pan C, Goldstein BA, Bernstein JA, Chaib H, Merker JD, Goldfeder RL, Enns GM, David SP (2014) Clinical interpretation and implications of whole-genome sequencing. JAMA 311:1035–1045
Roach JC, Glusman G, Smit AF, Huff CD, Hubley R, Shannon PT, Rowen L, Pant KP, Goodman N, Bamshad M (2010) Analysis of genetic inheritance in a family quartet by whole-genome sequencing. Science 328:636–639
Kubo Y, Sekiya S, Ohigashi M, Takenaka C, Tamura K, Nada S, Nishi T, Yamamoto A, Yamaguchi A (2005) ABCA5 resides in lysosomes, and ABCA5 knockout mice develop lysosomal disease-like symptoms. Mol Cell Biol 25:4138–4149
Petry F, Ritz V, Meineke C, Middel P, Kietzmann T, Schmitz-Salue C, Hirsch-Ernst KI (2006) Subcellular localization of rat Abca5, a rat ATP-binding-cassette transporter expressed in Leydig cells, and characterization of its splice variant apparently encoding a half-transporter. Biochem J 393:79–87
Patch A-M, Nones K, Kazakoff SH, Newell F, Wood S, Leonard C, Holmes O, Xu Q, Addala V, Creaney J (2018) Germline and somatic variant identification using BGISEQ-500 and HiSeq X Ten whole genome sequencing. PLoS ONE 13:e0190264
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760
Etherington GJ, Ramirez-Gonzalez RH, MacLean D (2015) bio-samtools 2: a package for analysis and visualization of sequence and alignment data with SAMtools in Ruby. Bioinformatics 31:2565–2567
Cibulskis K, McKenna A, Fennell T, Banks E, DePristo M, Getz G (2011) ContEst: estimating cross-contamination of human samples in next-generation sequencing data. Bioinformatics 27:2601–2602
Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, Jordan T, Shakir K, Roazen D, Thibault J (2013) From FastQ data to high-confidence variant calls: the genome analysis toolkit best practices pipeline. Curr protoc bioinformatics. 43:10–11
Larkin MA, Blackshields G, Brown N, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948
DeStefano GM, Kurban M, Anyane-Yeboa K, Dall'Armi C, Di Paolo G, Feenstra H, Silverberg N, Rohena L, López-Cepeda LD, Jobanputra V (2014) Mutations in the cholesterol transporter gene ABCA5 are associated with excessive hair overgrowth. PLoS Genet 10:e1004333
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E (2015) Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American college of medical genetics and genomics and the association for molecular pathology. Genet med 17:405
Kumar P, Henikoff S, Ng PC (2009) Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc 4:1073–1081
Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR (2010) A method and server for predicting damaging missense mutations. Nat Methods 7:248–249
Stenson PD, Mort M, Ball EV, Evans K, Hayden M, Heywood S, Hussain M, Phillips AD, Cooper DN (2017) The Human Gene Mutation Database: towards a comprehensive repository of inherited mutation data for medical research, genetic diagnosis and next-generation sequencing studies. Hum Genet 136:665–677
Marcus FI, McKenna WJ, Sherrill D, Basso C, Bauce B, Bluemke DA, Calkins H, Corrado D, Cox MG, Daubert JP (2010) Diagnosis of arrhythmogenic right ventricular cardiomyopathy/dysplasia: proposed modification of the task force criteria. Circulation 121:1533–1541
Yarlagadda RK, Iwai S, Stein KM, Markowitz SM, Shah BK, Cheung JW, Tan V, Lerman BB, Mittal S (2005) Reversal of cardiomyopathy in patients with repetitive monomorphic ventricular ectopy originating from the right ventricular outflow tract. Circulation 112:1092–1097
Komura S, Chinushi M, Furushima H, Hosaka Y, Izumi D, Iijima K, Watanabe H, Yagihara N, Aizawa Y (2010) Efficacy of procainamide and lidocaine in terminating sustained monomorphic ventricular tachycardia. Circ J 74:1003240657–1003240657
Haqqani HM, Kalman JM, Roberts-Thomson KC, Balasubramaniam RN, Rosso R, Snowdon RL, Sparks PB, Vohra JK, Morton JB (2009) Fundamental differences in electrophysiologic and electroanatomic substrate between ischemic cardiomyopathy patients with and without clinical ventricular tachycardia. J Am Coll Cardiol 54:166–173
Soejima K, Stevenson WG, Sapp JL, Selwyn AP, Couper G, Epstein LM (2004) Endocardial and epicardial radiofrequency ablation of ventricular tachycardia associated with dilated cardiomyopathy: the importance of low-voltage scars. J Am Coll Cardiol 43:1834–1842
Deal BJ, Miller SM, Scagliotti D, Prechel D, Gallastegui JL, Hariman R (1986) Ventricular tachycardia in a young population without overt heart disease. Circulation 73:1111–1118
Hasdemir C, Ulucan C, Yavuzgil O, Yuksel A, Kartal Y, Simsek E, Musayev O, Kayikcioglu M, Payzin S, Kultursay H (2011) Tachycardia-induced cardiomyopathy in patients with idiopathic ventricular arrhythmias: the incidence, clinical and electrophysiologic characteristics, and the predictors. J Cardiovasc Electrophysiol 22:663–668
Cano O, Hutchinson M, Lin D, Garcia F, Zado E, Bala R, Riley M, Cooper J, Dixit S, Gerstenfeld E (2009) Electroanatomic substrate and ablation outcome for suspected epicardial ventricular tachycardia in left ventricular nonischemic cardiomyopathy. J Am Coll Cardiol 54:799–808
Gilissen C, Hehir-Kwa JY, Thung DT, van de Vorst M, van Bon BW, Willemsen MH, Kwint M, Janssen IM, Hoischen A, Schenck A (2014) Genome sequencing identifies major causes of severe intellectual disability. Nature 511:344
Yuen RK, Thiruvahindrapuram B, Merico D, Walker S, Tammimies K, Hoang N, Chrysler C, Nalpathamkalam T, Pellecchia G, Liu Y (2015) Whole-genome sequencing of quartet families with autism spectrum disorder. Nat Med 21:185
Ye D, Meurs I, Ohigashi M, Calpe-Berdiel L, Habets KL, Zhao Y, Kubo Y, Yamaguchi A, Van Berkel TJ, Nishi T (2010) Macrophage ABCA5 deficiency influences cellular cholesterol efflux and increases susceptibility to atherosclerosis in female LDLr knockout mice. Biochem Biophys Res Commun 395:387–394
Fu Y, Hsiao J-HT, Paxinos G, Halliday GM, Kim WS (2015) ABCA5 regulates amyloid-β peptide production and is associated with Alzheimer's disease neuropathology. J Alzheimer's Dis 43:857–869
Goonasekara CL, Balse E, Hatem S, Steele DF, Fedida D (2010) Cholesterol and cardiac arrhythmias. Expert review of cardiovascular therapy 8:965–979
Spezzacatene A, Sinagra G, Merlo M, Barbati G, Graw SL, Brun F, Slavov D, Di Lenarda A, Salcedo EE, Towbin JA, Saffitz JE, Marcus FI, Zareba W, Taylor MR, Mestroni L, Familial Cardiomyopathy R (2015) Arrhythmogenic phenotype in dilated cardiomyopathy: natural history and predictors of life-threatening arrhythmias. J Am Heart Assoc 4:e002149
Hancarova M, Malikova M, Kotrova M, Drabova J, Trkova M, Sedlacek Z (2018) Association of 17q24. 2–q24. 3 deletions with recognizable phenotype and short telomeres. Am J Med Genet Part A 176:1438–1442
Stewart DR, Pemov A, Johnston JJ, Sapp JC, Yeager M, He J, Boland JF, Burdett L, Brown C, Gatti RA (2014) Dubowitz syndrome is a complex comprised of multiple, genetically distinct and phenotypically overlapping disorders. PLoS ONE 9:e98686
Vergult S, Dauber A, Delle Chiaie B, Van Oudenhove E, Simon M, Rihani A, Loeys B, Hirschhorn J, Pfotenhauer J, Phillips JA (2012) 17q24 2 microdeletions: a new syndromal entity with intellectual disability, truncal obesity, mood swings and hallucinations. Euro J Hum Genet 20:534
Acknowledgements
We are thankful to the proband and all the family members for participating in our study. We are thankful to the China National GeneBank and Shenzhen Peacock Plan (No. KQTD20150330171505310).
Funding
The National Natural Science Foundation, China (81570287, 81770315, 81770316) and Taishan Scholarship, Shandong, China (Silin Pan).
Author information
Authors and Affiliations
Contributions
SB and SP designed the study. Z conducted acquisition and analysis of all the clinical data. YH, PH, and YG made WES pipeline. SB, SK, YL, JL, and ZW analyzed the data. SB, SK, and YL wrote the manuscript. XL and XX supervised manuscript preparation and edited the manuscript.
Corresponding authors
Ethics declarations
Conflict of Interest
The authors confirm that there are no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Du, Z., Kuang, S., Li, Y. et al. Family-Based Whole Genome Sequencing Identified Novel Variants in ABCA5 Gene in a Patient with Idiopathic Ventricular Tachycardia. Pediatr Cardiol 41, 1783–1794 (2020). https://doi.org/10.1007/s00246-020-02446-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00246-020-02446-4