Abstract
Brucellosis is a notifiable disease induced by a facultative intracellular Brucella pathogen. In this study, eight Brucella abortus and eighteen Brucella melitensis strains from Egypt were annotated and compared with RB51 and REV1 vaccines respectively. RAST toolkit in the BV-BRC server was used for annotation, revealing genome length of 3,250,377 bp and 3,285,803 bp, 3289 and 3323 CDS, 48 and 49 tRNA genes, the same number of rRNA (3) genes, 583 and 586 hypothetical proteins, 2697 and 2726 functional proteins for B. abortus and B. melitensis respectively. B. abortus strains exhibit a similar number of candidate genes, while B. melitensis strains showed some differences, especially in the SRR19520422 Faiyum strain. Also, B. melitensis clarified differences in antimicrobial resistance genes (KatG, FabL, MtrA, MtrB, OxyR, and VanO-type) in SRR19520319 Faiyum and (Erm C and Tet K) in SRR19520422 Faiyum strain. Additionally, the whole genome phylogeny analysis proved that all B. abortus strains were related to vaccinated animals and all B. melitensis strains of Menoufia clustered together and closely related to Gharbia, Dameitta, and Kafr Elshiek. The Bowtie2 tool identified 338 (eight B. abortus) and 4271 (eighteen B. melitensis) single nucleotide polymorphisms (SNPs) along the genomes. These variants had been annotated according to type and impact. Moreover, thirty candidate genes were predicted and submitted at GenBank (24 in B. abortus) and (6 in B. melitensis). This study contributes significant insights into genetic variation, virulence factors, and vaccine-related associations of Brucella pathogens, enhancing our knowledge of brucellosis epidemiology and evolution in Egypt.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Brucella is a stealth pathogen that evades immune response and causes brucellosis which is regarded as a zoonotic infectious disease all over the world (Corbel 1997). The Brucella genus now includes twelve species that infect various domestic and wild-life animal species (Dawood et al. 2023). Six Brucella species have been identified as correlating to their pathogenicity and natural hosts. They are Brucella abortus (cattle), Brucella melitensis (sheep and goats), Brucella canis (dogs), Brucella suis (pigs), and Brucella neotomae (desert wood rat), Brucella ovis (rams) (Whatmore 2009; Blasco and Molina-Flores 2011). In Egypt, B. melitensis biovar 3 is the most common cause of animal and human infection, subsequent to B. abortus biovar 1 (Hamdy and Zaki 2018). Regardless of the implantation brucellosis control strategy in 1981, it remains widespread in Egypt (Hegazy et al. 2021).
Brucella’s genome is circular and very stable, and its size is 3.3 Mb (approximately 2.1 Mb on chromosome I and 1.2 Mb on chromosome II) (Michaux et al. 1993). There are striking similarities between B. abortus and B. melitensis corresponding to the homology degree found in DNA-DNA hybridization studies (Wang et al. 2012). Whole-genome sequencing (WGS) is regarded as the perfect technology for studying genomic variants of organisms in depth; it has become simple, cost-effective, and widely available (Tan et al. 2015).
Bioinformatics has significant roles not only in dealing with massive amounts of data, but also in the prediction, analysis, or interpretation of clinical and preclinical results, drug discovery, evaluation, and development (Khan et al. 2022). The comprehensive genome analysis provides detailed insights into genome quality evaluation, a phylogenetic tree, subsystem overview, antimicrobial resistance (AMR) genes, and candidate genes (Wang et al. 2020). Hence, in this study, we carried out detailed genetic characterization and comparative genomics analysis of some available Egyptian strains.
Materials & Methods
Retrieve Data
The sequences of B. abortus and B. melitensis strains in Egypt (from 2007 to 2019) were obtained from the National Center for Biotechnology Information (NCBI) repository and downloaded from European Nucleotide Archive (ENA) in fastq format (Tables S1 and S2). Most of the genomic analysis tools used in this study were done at the Bacterial and Viral Bioinformatics Resource Center (BV-BRC) server. Therefore, comparative genomics of available Egyptian strains (B. abortus n = 8 and B. melitensis n = 18) have been used in the current research to determine their closeness with vaccine strains (RB51 and Rev.1) for B. abortus and B. melitensis respectively.
Assembly and Annotation of Genomes
The processed fastq files are applied to the PATRIC assembly service which has seven different strategies. A Unicycler (V 0.4.8) pipeline used for the genome assembly of bacterial and archaeal genomes and the “AUTO” selection was applied to assemble the paired read to short read contigs. The unicycler employs the SPAdes de novo assembler to construct an initial assembly graph and subsequently refines by incorporating information from both short and long reads to generate high-quality, circular sequences and resolves complex repeat structures (Wick et al. 2017). A bandage program (V 0.8.1) was employed for visualization of these de novo assembly contigs. It primarily helps to identify errors and structure rearrangements in next-generation sequencing (NGS) data (Wick et al. 2015). Pilon (V 1.23) was used for correcting assembly errors for short reads to enhances the quality of draft genome assemblies producing more contiguous and accurate genome assemblies with better gene identification with remarkable accuracy in the identification of both small and large sequence variants (Walker et al. 2014). Additionally, TrimGalore was used for trimming the reads. It enables the automation of adapter trimming, and the removal of low-quality reads to enhance the quality of sequencing data. Also, SAMtools with default parameters were employed for reading mapping (Li et al. 2009). Finally, QUAST Version 5.0.2 is used for the automated and comprehensive analysis of genomics sequence assemblies to check the quality and completeness of the assembly output (Gurevich et al. 2013). Genome annotation refers to the process of locating functional components along the sequence of a genome. To annotate genomic characteristics in Brucella strains, rapid annotation using a subsystems technology tool kit (RASTtk) in BV-BRC was utilized (Aziz et al. 2008).
Comprehensive Genome Analysis
Comprehensive Genome Analysis was applied using the meta-service on the BV-BRC server that acquires raw data or the single or paired read to calculate genome assembly, annotation, quality control, AMR, and candidate genes that are specific for important functions (Davis et al. 2019).
Alignment Analysis
Progressive Mauve application was used to perform Multiple Sequence Alignment (MSA). Using MAUVE, the visually observable locally collinear blocks (LCBs), which are identified by specific color codes, were used to locate the genomic areas that were shared by all the matched sequences. LCBs indicate the homologous sequence found in two or more genomes without rearrangement (Darling et al. 2010). B. abortus RB51-AHVLA (Taxonomy ID: 1198700) and B. melitensis strain Rev.1 (Genome ID: 29459.409) were used as reference genomes from the PATRIC database. Moreover, at the level of Menoufia Governorate, Brucella from different centers was aligned with reference vaccine genome Rev.1.
Phylogenetic Analysis
Performing phylogenetic relationships in bacterial populations is critical to understanding the molecular evolutionary chronicle (O’Callaghan and Whatmore 2011). We used a BV-BRC phylogenetic tree called “Codon Trees” to know the evolutionary relationship between our strains and other outgroup strains to give an accurate view. It makes use of the predefined PATRIC protein global families (PGFams) (Davis et al. 2016), choosing 10–1000 single-copy families from among members of a genomic group. Alignments are produced for protein sequences of every family using Muscle (Edgar 2004), and for their corresponding nucleotide sequences, BioPython’s codon-align function was employed (Cock et al. 2009). To create confidence values, 100 rapid bootstrap rounds are executed within RaxML (Stamatakis et al. 2008).
Comparative Genomic Analysis
The comparative genome analysis was executed at the BV-BRC server. It combines two PATRIC tools (Protein Family Sorter and Comparative Pathways Viewer) as well as subsystems (Overbeek 2005; Overbeek et al. 2014). All three systems use two protein families: the local families (PLFams) for intra-genus comparisons and the global families (PGFams) for cross-genus comparisons (Davis et al. 2016). The PATRIC Protein Family Sorter tool is utilized to investigate the distribution patterns of protein families with different options: (1) present in all families to find the core genome, (2) absence from all families to examine the accessory genome and 3) mixed/either option which is the default is to determine the pangenome.
Variation Analysis
The variation analysis tool on the BV-BRC server finds the alterations from the reference (B. abortus RB51-AHVLA (Taxonomy ID: 1,198,700) and B. melitensis strain Rev.1 (Genome ID: 29,459.409)) and identifies single nucleotide variants (SNVs) and short insertions/deletions (indels) from aligned NGS data. Numerous tools have been developed such as BWA-mem and BWA-mem-strict (Li 2013), Bowtie2 (Langmead and Salzberg 2012), and LAST (Frith et al. 2010). FreeBayes (Garrison and Marth 2012) and SAMtools (Li et al. 2009) are the two SNP callers. Nevertheless, even minor variations could result in a significant number of variant calls across the entire genome. Therefore, it is crucial to select a reliable variant caller for both SNVs and indels and further evaluate and refine it to achieve maximum efficiency when analyzing the data. The analysis was done using the four different aligners.
Gene Prediction
The assembled contigs files generated from the previous steps from the BV-BRC website were subjected to various gene prediction servers: GeneMark.hmm prokaryotic (version 3.25), GeneMarkS-2 (Lomsadze et al. 2018), and EasyGene 1.2 Gene finding in prokaryotes (Larsen and Krogh 2003). To find the most concealed state sequence, the GeneMark.hmm server utilizes a hidden Markov model (HMM) architecture and the Viterbi algorithm, while GeneMarkS-2 is an ab initio technique that finds species-specific (native) genes using a model generated from self-training, as well as heuristic models meant to identify horizontal gene transfers. To evaluate GeneMarkS-2’s precision, the identified genes were confirmed through Clusters of Orthologous Groups (COG) proteomics experiments, annotation and N-terminal protein sequencing. EasyGene is also based on an algorithmic estimation of an HMM for an entirely new genome. After identifying the putative genes, they are evaluated using the HMM score and according to their scores and the length of the open reading frame (ORF), the statistical significance of the genes is calculated.
Molecular Evolutionary, Phylogenetic Analysis, and Conserved Regions of Predicted Genes
The 30 predicted genes underwent BLASTN and multiple sequence alignments from various genotypes via ClustalW multiple alignments impeded in BIOEDIT 7.2 software. The conserved regions refer to parts of DNA or protein sequences that have been preserved during evolution and contain functional elements such as protein-coding, non-coding regulatory domains, or structural motifs. In DNA sequences, the terms “similarity” and “identity” are used interchangeably to describe the degree of resemblance between two sequences, with identity specifically indicating that the two sequences are identical in that region. High sequence identity between regions of DNA typically indicates conservation, as these regions have been preserved across multiple organisms and may have important functional implications. These regions were identified by comparing sequences from multiple organisms or individuals. The “find conserved regions” option in the BIOEDIT 7.2 software with the default parameters of minimum length = 15 and max average entropy = 0.2. was applied to detect the conserved regions in the thirty predicted genes. These parameters ensure that identified regions meet a certain level of similarity and length to be considered conserved (Zhang et al. 2000).
After that, MEGA11 software and the ClustalW tool that impeded in it were employed for the phylogenetic analysis of the thirty predicted genes (Tamura et al. 2021). The multiple sequence alignment was performed using MAFFT server which offers various multiple alignment strategies. They are classified into three types, (a) the progressive method, (b) the iterative refinement method with the WSP score, and (c) the iterative refinement method using both the WSP and consistency scores. In order to obtain more accurate alignments, three new options, L-INS-i, G-INS-i, and E-INS-i, were used to align sequences globally using the Needleman-Wunsch algorithm. The five different constructed phylogenetic tree methods in MEGA11 software were used. Not only computing pairwise distances and overall mean distances but also different models were assessed by MEGA11 software. The best model selection “find best DNA/protein models” option was applied using MEGA to justify the choice of model for phylogenetics. Then the phylogenetic analysis was conducted on nucleotide substitution type and choice of the best model General Time Reversible Model (GTR). The GTR model is designed for inferring evolutionary relationships from DNA sequence data by accounting for different rates of nucleotide substitutions and the reversibility of these processes over time, making it an accurate tool for phylogenetic analysis. The initial tree(s) for the heuristic search were conducted automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using bootstrap methods, and the GTR model and then selecting the topology with superior log likelihood value. Phylogenetic trees use bootstrap values to measure the confidence level of sequence-based phylogeny on a scale of 1 to 100.
Results
Genome Assembly and Annotation
A comprehensive genome analysis of two Brucella (B.) species in Egypt was reported. The genome assembly of the eight B. abortus strains showed almost similar characteristics; contigs average were (30–35), genome length of 3,250,377 bp, and G + C content of 57.26%. The N50 length is 246,321 bp, which is recognized as the shortest sequence length at 50% of the genome. The L50 count was 6, which is known as the minimum number of contigs whose length summation yields N50 (Table 1). The B. melitensis strains were estimated with different numbers of contigs with an average of 26 in most of the strains (Dakahilia, Aswan, Ismailia, Beheira, and Beni-suef) and high numbers in (Asyut 40, Giza 42, and Beheira 58), an average of estimated genome length was 3,285,803 bp and G + C content of 57.25%. Also, the N50 length was 260,369 bp, and also the L50 was 4 (Table 2).
The genome annotation of Brucella species had an average of 3289 and 3323 coding DNA sequence (CDS), 48 and 49 transfer RNA genes (tRNA), 3 ribosomal RNA genes (rRNA), 583, 586 hypothetical proteins, and 2697, 2726 functional proteins in B. abortus and B. melitensis respectively (Fig. 1). These functional proteins estimated an average of 939 proteins have Enzyme Commission (EC) numbers, 805, 814 have Gene Ontology (GO), and 721, 731 proteins were linked to KEGG pathways in B. abortus and B. melitensis respectively. PATRIC annotation possessed 3190 and 3213 PLFams and 3220, and 3239 PGFams proteins for B. abortus and melitensis respectively (Tables 1 and 2).
The circular genome of Brucella (A) is illustrated, along with the circular genomes of eight B. abortus (B) and eighteen B. melitensis (C). The figure represents various genomic annotations, arranged from outside to inside, such as contigs, CDS on the forward and reverse strands, tRNA, CDS of AMR genes, CDS of virulence factor, GC content, and GC skew. The colors of CDS on the two strands reflect the subsystem to which these genes belong (see Subsystems below)
Candidate Genes and Antimicrobial Resistance Genes
Numerous annotated genes in B. abortus exhibit similarities with well-known genes associated with antibiotic resistance, transporters, drug targets as well as virulence factors, as indicated in (Table 3), while the AMR patterns in (Table 4) were consistent across all eight strains.
While the 18 B. melitensis strains showed some differences, especially in the SRR19520422 Faiyum strain in candidate genes (Table 5). Otherwise, SRR19520319 Faiyum and SRR19520422 Faiyum strains revealed differences in antimicrobial resistance genes comprising antibiotic activation enzyme (KatG), antibiotic target replacement protein FabL-like and regulator of antibiotic resistance gene expression (MtrA, MtrB, OxyR, VanO-type) for SRR19520319 Faiyum strain, and altering antibiotic target enzyme (Erm C) and an efflux pump that confers antibiotic resistance (Tet K) for SRR19520422 Faiyum strain (Table 4).
From the above results, the comprehensive genome analysis of the two species of Brucella and between each species and its strain provide striking resemblance in the genome and protein feature except slightly differences which proven the stability of Brucella species genome. However, the AMR genes profile displayed some novel genes that were not identified or with limited known to Brucella which may acquire from other species microorganisms.
Subsystems in B. abortus and B. melitensis
A subsystem is a collection of proteins that work together to carry out a certain biological process or structural complex. The pie chart summarizes the features of each subsystem and their coverage. A total of 83, 92 genes have been allocated to several subsystems, with the metabolism of amino acids receiving the most (37, 42). It was organized according to this pattern of Subsystem Counts (Subsystems, Genes), which means that in the case of B. abortus metabolism, it is a specific biological process with 96 subsystems controlled by 37 genes (Fig. 2).
Comparative Genome Analysis
Genome Alignment
Multiple sequence alignments of eight and eighteen genomes of B. abortus and B. melitensis with their reference vaccine genomes (RB51 and Rev.1) respectively were illustrated in (Figs. 3 and 4) using the progressive Mauve alignment program. Each genome is put out horizontally on a black horizontal center line that contains its name. Colored blocks represented homologous segments, while colored lines between genomes denote rearrangements or inversions (s-shape). Blocks positioned above the center line signify alignment in the forward direction of the initial genomic sequence, while those below indicate alignment in the reverse complement (inverse) direction. Moreover, at the level of Menoufia Governorate, Brucella strains from different centers are aligned with the reference vaccine genome Rev.1 illustrated in (Fig. 5).
The multiple sequence alignment of B. abortus is illustrated. The black circle represents the inversion region between two genomes of B. abortus, while colored lines between genomes in black boxes represent rearrangements of genomic sequences. The whole genome phylogeny (eight genomes) of B. abortus which was built on the protein and gene sequences for those 1000 genes performed at the PATRIC server confirmed the relatedness of B. abortus strains. The length of a branch is relative to the total number of changes at each site
The multiple sequence alignment of B. melitensis alignment is displayed. The black circles represent the inversion regions between the some of genomes of B. melitensis. The red box represented the same arrangement of the genomic sequences, while colored lines between genomes in the black box represented rearrangements of the genomic sequences. The phylogenetic tree for 18 whole genomes of B. melitensis which was built on the protein and gene sequences for those 1000 genes performed at PATRIC server confirmed the relatedness of B. melitensis strains. The length of a branch is relative to the total number of changes at each site (Color figure online)
The multiple sequence alignment of Brucella species is illustrated at Menoufia governorate centers. There is no rearrangement and inversion between the different B. melitensis species except for some slight inversions at the regions pointed by the arrow. The figure showed the conservation regions in different B. melitensis species at Menoufia centers. The phylogenetic tree for 22 whole genomes of Menoufia governorate from different centers which was built on the protein and gene sequences for those 1000 genes performed at the PATRIC server confirmed the close relatedness of B. melitensis strains. The length of a branch is relative to the total number of changes at each site
Phylogenetic Analysis
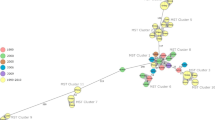
We constructed a whole genome phylogeny (90 genomes) involving Egyptian strains (n = 26) along with other available Egyptian and global strains. The tree was built on the protein and gene sequences for those 1000 genes. The whole genome phylogeny is clustered into six clades. The phylogeny indicated that all Egyptian B. abortus and B. melitensis strains are closely distance from each other. All B. melitensis strains of Menoufia clustered together and closely related to Gharbia, Dameitta, and Kafr Elshiek. On the opposite hand, B. melitensis stains of Giza were closely related to Beheira and Dakahlia. Also, B. abortus strains were related to vaccinated animals. B. Suis strains of Cairo were strictly related to B. Suis F7/06–2 from Germany. When compared to other B. abortus and B. melitensis strains, B. canis, B. ovis, B. Ceti, B. suis, B. neotomae, B. microti, and B. pinnipedialis strains were used as outgroups (Fig. 6).
The whole genome phylogeny (90 genomes) involving Egyptian strains (n = 26) with other available Egyptian and global strains is shown. The tree was built on the protein and gene sequences for those 1000 genes performed at the PATRIC server. Branch support values are obtained using 100 bootstrap and the length of a branch is relative to the total number of changes at each site
Core and Accessory Gene Compositions
Pangenome refers to the totality of shared and distinct genetic material present in a particular species that combines the genetic information of all the genomes sampled. It helps to better understand the comprehension composition and evolution of gene content within a population. The core genome is the identification of protein families that are either conserved across all the genomes, and that are conserved only in specific subsets of selected genomes are the accessory proteome.
In the case of B. abortus (8 genomes) and B. melitensis (18 genomes), the pan-genome analysis revealed 2677 and 2650 core genes and zero accessory genes of 2742 and 3911 gene pan-genome respectively. It is indicated that the genomes of these two Brucella species are highly similar, and they share most of their genetic material. The absence of accessory genes in the pan-genome analysis indicates that there is little genetic diversity among the sampled strains of these two species as shown in Fig. 6.
Single Nucleotide Polymorphism (SNPs)
Using the Bowtie2 tool identified 338 SNPs in all eight B. abortus genomes against RB51-AHVLA vaccine strain (reference genome) and from these SNPs 198 were commonly identified in all read libraries that we removed. The variants were annotated according to their possible effect and location on coding sequences (CDSs) via SnpEff (Cingolani et al. 2012). Variants type of classification exhibited 95 insertion variants, 28 nonsynonymous variants, 58 synonym variants, and 6 deletion variants. There were 26 frameshifts predicted, with 11 in-frame insertions 1 disrupted in-frame deletion, 37 intergenic regions, and 28 missense variants. Also, variants impact counted 53 modifiers, 40 moderate, 9 low, and 14 high.
In the case of B. melitensis, 4271 SNPs were identified in all eighteen genomes against the REV1 vaccine strain, and from these SNPs 2725 were commonly shared in all read libraries that we ignored. Also, variants type of classification revealed 394 insertion variants, 1831 nonsynonymous variants, 2229 synonym variants, and 200 deletion variants. Moreover, there were 230 frameshifts predicted, with 50 in-frame insertions, 20 in-frame deletions, 10 disrupted in-frame insertions, 5 disrupted in-frame deletions, and 773 intergenic regions, 1780 missense variants, 19 stop lost, 31 stop gained, 3 stop retained variants, 8 conservatives in-frame insertion, 8 conservatives in-frame deletion and 15 stop lost & splice region variants. In addition, variants impact counted 778 modifiers, 1850 moderate, 1126 low, and 228 high.
Conserved Regions, Molecular Evolutionary and Phylogenetic Analysis of Predicted Genes
Conserved Regions
The gene’s predicted servers (GeneMark.hmm prokaryotic (version 3.25), GeneMarkS-2, and EasyGene 1.2 Gene finding in prokaryote) were used to predict B. abortus (SRR12368034) and B. melitensis (SRR12368033). They revealed 1937 predicted genes for B. abortus and 1448 for B. melitensis. Hence, we submitted the most similar query genes to database sequence (24) for B. abortus and (6) for B. melitensis and accession numbers were obtained for the submitted genes. Using BIOEDIT for MSA, each gene has different numbers of conserved regions, segment length, and average entropy (Hx) (Tables 6 and 7).
Molecular Evolutionary
Maximum Likelihood Estimation of Substitution Matrix
The Tamura-Nei (1993) model was utilized to calculate substitution patterns and rates, where each element in the model reflects the probability of substitution (r) from one base to another (row to column). Transversional substitution rates were given in italics, while transitional substitution rates were shown in bold. For the sake of simplicity, the summation of r values is set to 100. A = 21.83%, T/U = 20.96%, C = 28.44%, and G = 28.78% are the nucleotide frequencies (Table S3). To estimate ML values, a tree topology was constructed spontaneously, and the log was -40,491.506. The study involved 30 nucleotide sequences, and gaps as well as incomplete data points were excluded using the full deletion option, resulting in a final dataset with 106 locations (Table S4).
Maximum Likelihood Estimation of Transition/Transversion Bias
The estimated Transition/Transversion bias (R) is 0.89. Each of the nucleotide frequencies is equal to 25.00%. To estimate ML values, a tree topology was instantly calculated, and the log was − 40,707.647.
Average Evolutionary Divergence Over All Sequence Pairs
The rejection odds of the null hypothesis, indicating whether sequences evolved with the same substitution pattern based on differences in base composition biases (Disparity Index test), were determined, and corresponding P-values below the diagonal were computed using a Monte Carlo test with 500 replicates. Significant P-values are less than 0.05 (highlighted in yellow). An assessment of the disparity index per site is presented for each sequence pair above the diagonal. The study involved 30 nucleotide sequences, with gaps and incomplete data points excluded using the full deletion option, resulting in a dataset of 106 points and an estimated distance of 5.37; the final dataset comprised a total of 4173 locations (Table S5).
Phylogenetic Analysis
MEGA11 software was used to analyze the 30 predicted genes using the Maximum Likelihood, Neighbor-joining technique, Minimum-Evolution, Unweighted Pair Group Mean Average “UPGMA” and Maximum Parsimony. Based on the lowest BIC scores (Bayesian Information Criterion), the General Time Reversible model (GTR) was found to be the best model for describing the substitution pattern. Each model’s AICc value (Akaike Information Criterion, corrected), Maximum Likelihood value (lnL), and number of parameters (including branch lengths) are also provided in (Table 8). To account for non-uniformity of evolutionary rates among sites, the discrete Gamma distribution with 5 rate categories (+ G) was used, as well as the assumption that a certain fraction of sites is evolutionarily invariable (+ I). If relevant, gamma shape parameter estimates and/or the fraction of invariant sites are also reported.
There were no discernible dissimilarities between the two methods. We conducted a phylogenetic analysis with both edited (Fig. 7) and unedited (Fig. 8) alignment and found no distinguishable differences between them. The phylogenetic tree and evolutionary relationships using these approaches were the same for the thirty genes of B. abortus and B. melitensis, therefore we justified with a maximum likelihood tree (Figs. 7 and 8) and Neighbour-joining tree (Fig. S1). According to the submitted genes, the two Brucella species (abortus and melitensis) were closely related to each other.
The molecular evolution was assumed using the Maximum Likelihood approach General Time Reversible model. This is the tree with the highest log likelihood (− 40,491.51). Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the Maximum Composite Likelihood (MCL) approach and then selecting the topology with superior log likelihood value. This analysis involved 30 nucleotide sequences. Codon positions included were 1st + 2nd + 3rd + Noncoding. There were a total of 1081 positions in the final dataset
The evolutionary history was inferred by using the Maximum Likelihood method and General Time Reversible model. The tree with the highest log likelihood (− 40,549.08) is shown. The percentage of trees in which the associated taxa clustered together is shown below the branches. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the Maximum Composite Likelihood (MCL) approach and then selecting the topology with superior log likelihood value. This analysis involved 30 nucleotide sequences. Codon positions included were 1st + 2nd + 3rd + Noncoding. There was a total of 4173 positions in the final dataset
Discussion
Brucellosis is often considered one of the world’s most serious zoonotic diseases. It causes enormous economic losses in both developed and developing countries. Despite massive attempts to eradicate Brucella in many countries of the world, this disease has disseminated worldwide (Elrashedy et al. 2022). The complete genome of Brucella is 3.3 Mb (approximately 2.1 Mb on chromosome I and 1.2 Mb on chromosome II) (Michaux et al. 1993). Genomic annotation and comparative genome analysis of Brucella species are important to reveal the genetic features and get a deep look at the genetic variation in the form of SNPs for validating the vaccine strain. In this study, B. abortus strains annotation displayed striking similarities in a variety of features to those previously reported in studies (Wang et al. 2020; Yu et al. 2015). On the other hand, annotation of B. melitensis strains was agreed upon in a study performed by (Karthik et al. 2021) and this may be due to the genome stability of Brucella this study’s findings confirm that there are significant resemblances between the two Brucella species according to the homology degree.
In this study, there are 33 antimicrobial resistance genes identified by the PATRIC server in the two brucella species. The intracellular lifestyle of brucellae, which prevents the penetration of certain antimicrobials into the cells, may have a role in the emergence of resistance in these bacteria. As a result, resistance and virulence mechanisms at the genomic, proteomic, and transcriptome levels trigger the alert about an increase in the number of residence genes. The stealthy Brucella pathogen has different AMR mechanisms such as enzymatic activation and inactivation of the antimicrobial agent, efflux pumps, replacement of protein or alteration occurring in cell wall charge via protein, drug target modification and resistance genes regulating the gene expression (Biswas et al. 2008; Dadar et al. 2023). DNA gyrase enzyme (gyrA and gyrB) has the primary role in DNA supercoiling, and inhibition of this activity using quinolones or fluoroquinolones generation is combined with rapid killing of the pathogenic Brucella (Spencer and Panda 2023). However, rifampicin is composed of four distinguishing subunits (rpoA, rpoB, rpoC and rpoD) and it has a mode of action that inhibits the DNA-dependent RNA polymerase, and its resistance is caused by the mutation at 81 bp in the rpoB gene (Portelli et al. 2020). Also, FosX is an AMR gene that can inactivate Fosfomycin (Bolotin et al. 2021).
Furthermore, efflux pumps can be categorized into six groups, namely, the ATP-binding cassette (ABC), resistance nodulation cell division (RND), major facilitator superfamily (MFS), multi-antimicrobial and toxic compound extrusion (MATE), proteobacterial antimicrobial compound efflux (PACE) and small multidrug resistance (SMR) family (Du et al. 2018). In this study, efflux pumps in two families (ABC and RND) were accompanied by antibiotic resistance. MacAB (macrolide-specific ABC-type efflux carrier) involves macA and macB genes that perform resistance against macrolides composed of 14- and 15-membered compounds (Greene et al. 2018). The RND family is another efflux pump that features the triclosan-specific efflux protein TriABC-OpmH. In Pseudomonas aeruginosa, this protein comprises two membrane fusion proteins, TriA and TriB (Fabre et al. 2021). Although its role in Brucella is not yet understood, increased resistance to ciprofloxacin and imipenem suggests that the efflux pump in Brucella may provide quinolone resistance.
What’s more, folic acid (vitamin B9) is an important amino acid that enters the folate synthesis process which is vital for the replication of DNA. This process can be blocked by trimethoprim and sulfamethoxazole. The presence or absence of the folA (dihydrofolate reductase) or the folP (dihydropteroate synthase) can produce resistance to trimethoprim and sulfamethoxazole respectively (Biswas et al. 2008; Nurjadi et al. 2021). Additionally, gidB gene encodes a tRNA uridine 5-carboxymethyl aminomethyl modification enzyme that participates in post-transcriptional tRNA modifications. The gidB exhibited a decreased susceptibility to aminoglycoside antibiotics like gentamicin and streptomycin (Mikheil et al. 2012).
In Brucella, OxyR is a transcription factor that affects the bacteria’s ability to combat oxidative stress and resist reactive oxygen species (ROS) produced within the host cell. This protein controls the expression of different cellular components responsible for antioxidant defense mechanisms. It can trigger the activation of numerous genes, including catalase-peroxidase, superoxide dismutase, and other detoxification genes, which aid in the bacteria’s protection against ROS-induced damage (Kim and Mayfield 2000; Wang et al. 2022). Moreover, PgsA and GdpD contribute to the lipopolysaccharide (LPS) that has a very important role in Brucella virulence and survival (Elrashedy et al. 2022). In addition, these genes Alr, Ddl, S10p, S12p, EF-G, inhA, EF-Tu, Iso-tRNA, Dfr, kasA, rho, MurA, and fabI have been implicated in various cellular functions and pathways in Brucella. While some of these genes have been implicated in antibiotic resistance in other bacteria, their role in Brucella antibiotic resistance may not be extensively studied or well-established.
In this study, there are some differences in Fayioum strains of B. meliteinsis revealed in antimicrobial resistance genes which can be explained on the basis that Fayioum main raising cattle is beef collected from different sources (local, European, and Latin) with no vaccination representing a pool for mixing different Brucella strains from different sources. KatG is a gene that encodes for the catalase-peroxidase enzyme. This enzyme is responsible for the detoxification of ROS within bacterial cells and is therefore considered to play a key role in bacterial defense mechanisms, including resistance to antibiotics. Although KatG is not typically considered as a primary antibiotic resistance gene in Brucella, studies have shown that mutations in this gene can result in resistance to isoniazid, an anti-tuberculosis drug that is also used to treat brucellosis (Bollela et al. 2016). Additionally, The Erm(C) gene is a ribosomal RNA methylase gene that confers resistance to macrolide-lacosamide-streptogramin B (MLSB) antibiotics, including erythromycin, clindamycin, and streptogramin B (Khodabandeh et al. 2019). From the above mentioned, antibiotic-resistance genes have the potential to spread between different bacterial species through horizontal gene transfer, leading to the emergence of multidrug-resistant strains that are difficult to treat. Hence, it is crucial to monitor antibiotic resistance patterns across various bacterial species and develop suitable treatment strategies to ensure the effective management of bacterial infections.
In this study, whole genome phylogeny analysis for all Brucella strains was classified into six clades: B. melitensis clade; B. abortus clade; B. canis clade; B. suis clade; B. microti; B. pinnipedialis; B. ceti clade; and B. ovis clade. The evolutionary relationship constructed here fits with previous results (Azam et al. 2016). The transition mutations occur more frequently than transversion mutations and could have an impact on the evolution of protein-coding genes. This is because the DNA sequence undergoes less significant changes during transition mutations, which may result in fewer negative consequences on the protein-coding sequence. As a result, certain parts of the genome could experience higher rates of transition substitutions, possibly due to the less deleterious impact of these mutations on protein function in those regions.
By analyzing the pan-genome, we can gain a comprehensive understanding of the gene content and genetic evolution within a population, which provides valuable insights. Additionally, identifying core and accessory genes through pan-genome analysis can help us to better comprehend the genetic relatedness and variability of different strains of a particular species. When we apply this mechanism to B. abortus and B. melitensis, it suggests that these two species have a significant genetic resemblance, which is also supported by the absence of accessory genes in the sampled strains. In this study, the results are in agreement with (Wattam et al. 2014) who reported 2285 core genes in the Brucella species. Finally, This study enhances our knowledge of the epidemiology and evolution of brucellosis in Egypt by providing significant insights into the genetic diversity, potential virulence factors, and associations related to vaccines of Brucella pathogens.
Conclusion
Our investigation into Brucellosis involves the genomic analysis of eight B. abortus and eighteen B. melitensis strains from Egypt have yielded significant findings. The comprehensive comparison with RB51 and REV1 vaccines, annotation of genome characteristics, identification of antimicrobial resistance genes (AMR), and elucidation of phylogenetic relationships offer an understanding of the genetic landscape of this Brucella pathogen. The detection of AMR in two Brucella species indicates that Brucellae possess several strategies to withstand antimicrobial agents. One of these strategies is the intracellular lifestyle of Brucella, which hinders the entry of specific antimicrobials into the cells, contributing to the development of resistance. Also in this study, we predicted and submitted thirty candidate genes to GenBank. Interestingly, the distinct features observed in B. melitensis strains, particularly in the Faiyum isolate, highlight potential variations in virulence and resistance profiles. The clustering patterns in the whole genome phylogeny further emphasize the geographical relevance, linking B. abortus strains to vaccinated animals and grouping B. melitensis strains from Menoufia, Gharbia, Dameitta, and Kafr Elshiek. The detection of numerous SNPs and subsequent annotation enhances our knowledge of the molecular intricacies of the Brucella pathogen. This research increases our understanding of the epidemiology and evolution of brucellosis in Egypt by providing useful insights into the genetic diversity, putative virulence factors, and vaccine-related associations of these Brucella pathogens.
Data Availability
All the primary data used in the study were included in the article (and its supplementary files).
References
Azam S, Rao SB, Jakka P et al (2016) Genetic characterization and comparative genome analysis of Brucella melitensis isolates from India. Int J Genomics 2016:1–13. https://doi.org/10.1155/2016/3034756
Aziz RK, Bartels D, Best AA et al (2008) The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. https://doi.org/10.1186/1471-2164-9-75
Biswas S, Raoult D, Rolain J-M (2008) A bioinformatic approach to understanding antibiotic resistance in intracellular bacteria through whole genome analysis. Int J Antimicrob Agents 32:207–220. https://doi.org/10.1016/j.ijantimicag.2008.03.017
Blasco JM, Molina-Flores B (2011) Control and Eradication of Brucella melitensis Infection in Sheep and Goats. Vet. Clin. North Am. - Food Anim. Pract. 27
Bollela VR, Namburete EI, Feliciano CS et al (2016) Detection of katG and inhA mutations to guide isoniazid and ethionamide use for drug-resistant tuberculosis. Int J Tuberc Lung Dis 20:1099–1104. https://doi.org/10.5588/ijtld.15.0864
Bolotin V, Kovalenko G, Marchenko N et al (2021) Complete genome sequence of Brucella abortus 68, isolated from aborted fetal sheep in Ukraine. Microbiol Resour Announc. https://doi.org/10.1128/MRA.01436-20
Cingolani P, Platts A, Wang LL et al (2012) A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff. Fly (austin) 6:80–92. https://doi.org/10.4161/fly.19695
Cock PJA, Antao T, Chang JT et al (2009) Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 25:1422–1423. https://doi.org/10.1093/bioinformatics/btp163
Corbel M (1997) Brucellosis: an overview. Emerg Infect Dis 3:213–221. https://doi.org/10.3201/eid0302.970219
Dadar M, Brangsch H, Alamian S et al (2023) Whole-genome sequencing for genetic diversity analysis of Iranian Brucella spp. isolated from humans and livestock. One Heal 16:100483. https://doi.org/10.1016/J.ONEHLT.2023.100483
Darling AE, Mau B, Perna NT (2010) progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE 5:e11147. https://doi.org/10.1371/journal.pone.0011147
Davis JJ, Gerdes S, Olsen GJ et al (2016) PATtyFams: protein families for the microbial genomes in the PATRIC database. Front Microbiol. https://doi.org/10.3389/fmicb.2016.00118
Davis JJ, Wattam AR, Aziz RK et al (2019) The PATRIC bioinformatics resource center: expanding data and analysis capabilities. Nucleic Acids Res. https://doi.org/10.1093/nar/gkz943
Dawood AS, Elrashedy A, Nayel M et al (2023) Brucellae as resilient intracellular pathogens: epidemiology, host–pathogen interaction, recent genomics and proteomics approaches, and future perspectives. Front Vet Sci. https://doi.org/10.3389/fvets.2023.1255239
Du D, Wang-Kan X, Neuberger A et al (2018) Author correction: multidrug efflux pumps: structure, function and regulation. Nat Rev Microbiol 16:577–577. https://doi.org/10.1038/s41579-018-0060-x
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. https://doi.org/10.1093/nar/gkh340
Elrashedy A, Gaafar M, Mousa W et al (2022) Immune response and recent advances in diagnosis and control of brucellosis. Ger J Vet Res 2:10–24. https://doi.org/10.51585/gjvr.2022.1.0033
Fabre L, Ntreh AT, Yazidi A et al (2021) A “drug sweeping” state of the TriABC Triclosan efflux pump from pseudomonas aeruginosa. Structure 29:261-274.e6. https://doi.org/10.1016/j.str.2020.09.001
Frith MC, Wan R, Horton P (2010) Incorporating sequence quality data into alignment improves DNA read mapping. Nucleic Acids Res 38:e100–e100. https://doi.org/10.1093/nar/gkq010
Garrison E, Marth G (2012) Haplotype-based variant detection from short-read sequencing, pp. 1–9
Greene NP, Kaplan E, Crow A, Koronakis V (2018) Antibiotic resistance mediated by the MacB ABC transporter family: a structural and functional perspective. Front Microbiol. https://doi.org/10.3389/fmicb.2018.00950
Gurevich A, Saveliev V, Vyahhi N, Tesler G (2013) QUAST: quality assessment tool for genome assemblies. Bioinformatics 29:1072–1075. https://doi.org/10.1093/bioinformatics/btt086
Hamdy MER, Zaki HM (2018) Detection of virulence-associated genes in Brucella melitensis biovar 3, the prevalent field strain in different animal species in Egypt. Open Vet J 8:112. https://doi.org/10.4314/OVJ.V8I1.17
Hegazy YM, Schley D, Ridler A et al (2021) Control of Brucella melitensis in endemic settings: a simulation study in the Nile Delta, Egypt. Transbound Emerg Dis 68:2364–2375. https://doi.org/10.1111/TBED.13897
Karthik K, Anbazhagan S, Thomas P et al (2021) Genome sequencing and comparative genomics of indian isolates of Brucella melitensis. Front Microbiol. https://doi.org/10.3389/FMICB.2021.698069
Khan K, Alhar MSO, Abbas MN et al (2022) Integrated bioinformatics-based subtractive genomics approach to decipher the therapeutic drug target and its possible intervention against brucellosis. Bioengineering 9:633. https://doi.org/10.3390/BIOENGINEERING9110633/S1
Khodabandeh M, Mohammadi M, Abdolsalehi MR et al (2019) Analysis of resistance to macrolide–lincosamide–streptogramin b among meca-positive Staphylococcus aureus isolates. Osong Public Heal Res Perspect 10:25–31. https://doi.org/10.24171/j.phrp.2019.10.1.06
Kim J, Mayfield J (2000) Identification of Brucella abortus OxyR and its role in control of catalase expression. J Bacteriol 182:5631–5633. https://doi.org/10.1128/JB.182.19.5631-5633.2000
Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. https://doi.org/10.1038/nmeth.1923
Larsen TS, Krogh A (2003) EasyGene–a prokaryotic gene finder that ranks ORFs by statistical significance. BMC Bioinformatics. https://doi.org/10.1186/1471-2105-4-21
Li H, Handsaker B, Wysoker A et al (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. https://doi.org/10.1093/bioinformatics/btp352
Li H (2013) Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. 00:1–3
Lomsadze A, Gemayel K, Tang S, Borodovsky M (2018) Modeling leaderless transcription and atypical genes results in more accurate gene prediction in prokaryotes. Genome Res 28:1079–1089. https://doi.org/10.1101/GR.230615.117
Michaux S, Paillisson J, Carles-Nurit MJ et al (1993) Presence of two independent chromosomes in the Brucella melitensis 16M genome. J Bacteriol 175:701–705. https://doi.org/10.1128/jb.175.3.701-705.1993
Mikheil DM, Shippy DC, Eakley NM et al (2012) Deletion of gene encoding methyltransferase (gidB) confers high-level antimicrobial resistance in Salmonella. J Antibiot (tokyo) 65:185–192. https://doi.org/10.1038/ja.2012.5
Nurjadi D, Zizmann E, Chanthalangsy Q et al (2021) Integrative analysis of whole genome sequencing and phenotypic resistance toward prediction of trimethoprim-sulfamethoxazole resistance in Staphylococcus aureus. Front Microbiol. https://doi.org/10.3389/fmicb.2020.607842
O’Callaghan D, Whatmore AM (2011) Brucella genomics as we enter the multi-genome era. Brief Funct Genomics 10:334–341. https://doi.org/10.1093/bfgp/elr026
Overbeek R (2005) The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res 33:5691–5702. https://doi.org/10.1093/nar/gki866
Overbeek R, Olson R, Pusch GD et al (2014) The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res 42:D206–D214. https://doi.org/10.1093/nar/gkt1226
Portelli S, Myung Y, Furnham N et al (2020) Prediction of rifampicin resistance beyond the RRDR using structure-based machine learning approaches. Sci Rep 10:18120. https://doi.org/10.1038/s41598-020-74648-y
Spencer AC, Panda SS (2023) DNA gyrase as a target for quinolones. Biomedicines 11:371. https://doi.org/10.3390/biomedicines11020371
Stamatakis A, Hoover P, Rougemont J (2008) A rapid bootstrap algorithm for the RAxML Web servers. Syst Biol 57:758–771. https://doi.org/10.1080/10635150802429642
Tamura K, Stecher G, Kumar S (2021) MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol 38:3022–3027. https://doi.org/10.1093/molbev/msab120
Tan K-K, Tan Y-C, Chang L-Y et al (2015) Full genome SNP-based phylogenetic analysis reveals the origin and global spread of Brucella melitensis. BMC Genomics 16:93. https://doi.org/10.1186/s12864-015-1294-x
Walker BJ, Abeel T, Shea T et al (2014) Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 9:e112963. https://doi.org/10.1371/journal.pone.0112963
Wang Y, Ke Y, Wang Z et al (2012) Genome sequences of three live attenuated vaccine strains of Brucella Species and implications for pathogenesis and differential diagnosis. J Bacteriol 194:6012–6013. https://doi.org/10.1128/JB.01483-12
Wang S, Wang W, Sun K et al (2020) Comparative genomic analysis between newly sequenced Brucella abortus vaccine strain A19 and another Brucella abortus vaccine S19. Genomics 112:1444–1453. https://doi.org/10.1016/J.YGENO.2019.08.015
Wang J, Liu J, Zhao Y et al (2022) OxyR contributes to virulence of Acidovorax citrulli by regulating anti-oxidative stress and expression of flagellin FliC and type IV pili PilA. Front Microbiol. https://doi.org/10.3389/fmicb.2022.977281
Wattam AR, Foster JT, Mane SP et al (2014) Comparative phylogenomics and evolution of the Brucellae reveal a path to virulence. J Bacteriol 196:920–930. https://doi.org/10.1128/JB.01091-13
Whatmore AM (2009) Current understanding of the genetic diversity of Brucella, an expanding genus of zoonotic pathogens. Infect Genet Evol 9:1168–1184. https://doi.org/10.1016/J.MEEGID.2009.07.001
Wick RR, Schultz MB, Zobel J, Holt KE (2015) Bandage: interactive visualization of de novo genome assemblies. Bioinformatics 31:3350–3352. https://doi.org/10.1093/bioinformatics/btv383
Wick RR, Judd LM, Gorrie CL, Holt KE (2017) Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLOS Comput Biol 13:e1005595. https://doi.org/10.1371/journal.pcbi.1005595
Yu D, Hui Y, Zai X et al (2015) Comparative genomic analysis of Brucella abortus vaccine strain 104M reveals a set of candidate genes associated with its virulence attenuation. Virulence 6:745–754. https://doi.org/10.1080/21505594.2015.1038015/SUPPL_FILE/KVIR_A_1038015_SM6583.XLS
Zhang Z, Schwartz S, Wagner L, Miller W (2000) A greedy algorithm for aligning DNA sequences. J Comput Biol 7:203–214. https://doi.org/10.1089/10665270050081478
Acknowledgements
The authors thank the Academy of Scientific Research and Technology (Scientists for Next Generation, cycle 7).
Funding
Open access funding provided by The Science, Technology & Innovation Funding Authority (STDF) in cooperation with The Egyptian Knowledge Bank (EKB). This study was supported by the Academy of Scientific Research and Technology (Scientists for Next Generation, cycle 7).
Author information
Authors and Affiliations
Contributions
AE and MEH led work on this study, wrote the main manuscript text, and prepared all the figures. MEH contributed to the investigations. NRA delved deeper into the analysis result and discussion. MN, AS and AZ supervised and contributed to the conceptualization of the study. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interest
The authors declare that they have no competing interests.
Ethical Approval
This research complies with relevant institutional, national, and international guidelines and legislation. This research does not include any studies with human participants or animals performed by authors.
Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Additional information
Handling editor: Ananias Escalante.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Elrashedy, A., Nayel, M., Salama, A. et al. Phylogenetic Analysis and Comparative Genomics of Brucella abortus and Brucella melitensis Strains in Egypt. J Mol Evol 92, 338–357 (2024). https://doi.org/10.1007/s00239-024-10173-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-024-10173-0