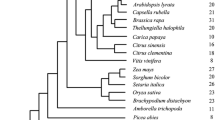
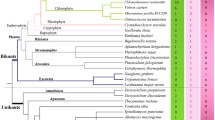
Abstract
HUA ENHANCER 1 (HEN1) is a pivotal mediator in protecting sRNAs from 3′-end uridylation and 3′ to 5′ exonuclease-mediated degradation in plants. Here, we investigated the pattern of the HEN1 protein family evolutionary history and possible relationships in the plant lineages using protein sequence analyses and conserved motifs composition, functional domain identification, architecture, and phylogenetic tree reconstruction and evolutionary history inference. According to our results, HEN1 protein sequences bear several highly conserved motifs in plant species retained during the evolution from their ancestor. However, several motifs are present only in Gymnosperms and Angiosperms. A similar trend showed for their domain architecture. At the same time, phylogenetic analysis revealed the grouping of the HEN1 proteins in the three main super clads. In addition, the Neighbor-net network analysis result provides some nodes have multiple parents indicating a few conflicting signals in the data, which is not the consequence of sampling error, the effect of the selected model, or the estimation method. By reconciling the protein and species tree, we considered the gene duplications in several given species and found 170 duplication events in the evolution of HEN1 in the plant lineages. According to our analysis, the main HEN1 superclass mostly showed orthologous sequences that illustrate the vertically transmitting of HEN1 to the main lines. However, in both orthologous and paralogs, we predicted insignificant structural deviations. Our analysis implies that small local structural changes that occur continuously during the folds can moderate the changes created in the sequence. According to our results, we proposed a hypothetical model and evolutionary trajectory for the HEN1 protein family in the plant kingdom.






Similar content being viewed by others
References
Adams KL, Cronn R, Percifield R, Wendel JF (2003) Genes duplicated by polyploidy show unequal contributions to the transcriptome and organ-specific reciprocal silencing. PNAS 100:8. https://doi.org/10.1073/pnas.0630618100
Bagowski CP, Bruins W, te Velthuis AJW (2010) The nature of protein domain evolution: shaping the interaction network. Curr Genomics 11:368–376
Bailey TL, Elkan C (1994) Fitting a mixture model by expectation maximization to discover motifs in biopolymers, In: Proceedings of the second international conference on intelligent systems for molecular biology, pp 28–36
Baranauske S, Mickute M, Plotnikova A et al (2015) Functional mapping of the plant small RNA methyltransferase: HEN1 physically interacts with HYL1 and DICER-LIKE 1 proteins. Nucleic Acids Res. https://doi.org/10.1093/nar/gkv102
Borges F, Martienssen R (2015) The expanding world of small RNAs in plants. Nat Rev Mol Cell Biol 16:727–741. https://doi.org/10.1038/nrm4085
Bryant D, Moulton V (2004) Neighbor-net: an agglomerative method for the construction of phylogenetic networks. Mol Biol Evol 21(2):255–265. https://doi.org/10.1093/molbev/msh018
Chen X (2009) Small RNAs and their roles in plant development. Annu Rev Cell Dev Biol 25:21–44. https://doi.org/10.1146/annurev.cellbio.042308.113417
De-Castro E, Sigrist CJA, Gattiker A et al (2006) ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res 34((Web Server Issue)):W362–W365
Di-Tommaso P, Moretti S, Xenarios I et al (2011) T-Coffee: a web server for the multiple sequence alignment of protein and RNA sequences using structural information and homology extension. Nucleic Acids Res 39((Web Server Issue)):W13–W17. https://doi.org/10.1093/nar/gkr245
Edgar CR (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32(5):1792–1797. https://doi.org/10.1093/nar/gkh340
Farrokhi N, Hajieghrari B (2020) Chronicles of dolos and apate in plant microRNAs. Biologia 75(12):2465–2465. https://doi.org/10.2478/s11756-020-00545-4
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Finn RD, Clements J, Eddy SR (2011) HMMER web server: interactive sequence similarity searching. Nucleic Acids Res 39:W29–W37. https://doi.org/10.1093/nar/gkr367
Frickey T, Lupas A (2004) CLANS: a Java application for visualizing protein families based on pairwise similarity. Bioinformatics 20(18):3702–3704. https://doi.org/10.1093/bioinformatics/bth444
Gabler F, Nam S, Till S et al (2020) Protein Sequence Analysis Using the MPI Bioinformatics Toolkit. Current Protocols in Bioinformatics 72:e108. https://doi.org/10.1002/cpbi.108
Gu Z, Steinmetz LM, Gu X et al (2003) Role of duplicate genes in genetic robustness against null mutations. Nature 421(6918):63–66. https://doi.org/10.1038/nature01198
Hajieghrari B, Farrokhi N (2021) Investigation on the conserved microRNA genes in higher plants. Plant Mol Biol Rep 39(1):10–23. https://doi.org/10.1007/s11105-020-01228-9
Hajieghrari B, Farrokhi N (2022) Plant RNA-mediated gene regulatory network. Genomics 114(1):409–442. https://doi.org/10.1016/j.ygeno.2021.12.020
Hajieghrari B, Farrokhi N, Goliaei B, Kavousi K (2019) The role of MicroRNAs in defense against viral phytopathogens. Physiol Mol Plant Pathol 107:8–13. https://doi.org/10.1016/j.pmpp.2019.04.008
Holm L (2020) Using Dali for protein structure comparison. Methods Mol Biol 2112:29–42. https://doi.org/10.1007/978-1-0716-0270-6_3
Huang Y, Ji L, Huang Q et al (2009) Structural insights into mechanisms of the small RNA methyltransferase HEN1. Nature 461:823–829. https://doi.org/10.1038/nature08433
Huson DH, Bryant D (2006) Application of phylogenetic networks in evolutionary studies. Mol Biol Evol 23(2):254–267. https://doi.org/10.1093/molbev/msj030
Ji L, Chen X (2012) Regulation of small RNA stability: methylation and beyond. Cell Res 22:624–636. https://doi.org/10.1038/cr.2012.36
Katoh K, Misawa K, Kuma K, Miyata T (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30(14):3059–3066. https://doi.org/10.1093/nar/gkf436
Kumar S, Filipski A (2007) Multiple sequence alignment: In pursuit of homologous DNA positions. Genomics Res 17:127–135. https://doi.org/10.1101/gr.5232407
Kurek I, Aviezer K, Erel N et al (1999) The wheat peptidyl prolyl cis-trans-isomerase FKBP77 is heat induced and developmentally regulated. Plant Physiol 119(2):693–704. https://doi.org/10.1104/pp.119.2.693
Lassmann T, Sonnhammer EL (2005) Kalign – an accurate and fast multiple sequence alignment algorithm. BMC Bioinform 6:298. https://doi.org/10.1186/1471-2105-6-298
Letunic I, Bork P (2021) Interactive tree of life (iTOL) v5; an online tool for phylogenetic tree display and annotation. Nucleic Acids Res 49(W1):W293–W296. https://doi.org/10.1093/nar/gkab301
Li J, Yang Z, Yu B et al (2005) Methylation protects miRNAs and siRNAs from a 3’-end uridylation activity in Arabidopsis. Curr Biol 15(16):1501–1507. https://doi.org/10.1016/j.cub.2005.07.029
Li F, Fan G, Lu C et al (2015) Genome sequence of cultivated Upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nat Biotechnol 33:524–530. https://doi.org/10.1038/nbt.3208
Mistry J, Chuguransky S, Williams L (2020) Pfam: the protein families database in 2021. Nucleic Acids Res. https://doi.org/10.1093/nar/gkaa913
Nguyen Ba AN, Pogoutse A, Provart N, Moses AM (2009) NLStradamus: a simple Hidden Markov Model for nuclear localization signal prediction. BMC Bioinform 10(1):202. https://doi.org/10.1186/1471-2105-10-202
Ohno S (1970) Evolution by gene duplication. Springer-Verlag, Heidelberg
Ramachandran V, Chen X (2008) Degradation of micro RNAs by a family of exoribonuclease in Arabidopsis. Science 321:1490–1492. https://doi.org/10.1126/science.1163728
Sanei M, Chen X (2015) Mechanisms of microRNA turnover. Curr Opin Plant Biol 27:199–206. https://doi.org/10.1016/j.pbi.2015.07.008
Schultz J, Copley RR, Doerks T et al (2000) SMART: a web-based tool for the study of genetically mobile domains. Nucleic Acids Res 28(1):231–234. https://doi.org/10.1093/nar/28.1.231
Sievers F, Higgins DG (2017) Clustal Omega for making accurate alignments of many protein sequences. Protein Sci 27(1):135–145. https://doi.org/10.1002/pro.3290
Singh A, Gautam V, Singh S et al (2018) Plant small RNAs: advancement in the understanding of biogenesis and role in plant development. Planta 248(3):545–558. https://doi.org/10.1007/s00425-018-2927-5
Tamura K, Stecher G, Kumar S (2021) MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol 38(7):3022–3027. https://doi.org/10.1093/molbev/msab120
Tkaczuk K L, Obarska A, Bujnicki J M (2006) Molecular phylogenetics and comparative modeling of HEN1, a methyltransferase involved in plant microRNA biogenesis. BMC Evol Biol 6(6). https://doi.org/10.1186/1471-2148-6-6
Wagner A (2005) Energy constraints on the evolution of gene expression. Mol Biol Evol 22(6):1365–1374. https://doi.org/10.1093/molbev/msi126
Wang X, Wang Y, Dou Y et al (2018) Degradation of unmethylated miRNA/miRNA*s by a DEDDy-type 3′ to 5′ exoribonuclease Artimmer 2 in Arabidopsis. PNAS 115:E6659–E6667. https://doi.org/10.1073/pnas.1721917115
Webb B, Sali A (2016) Comparative protein structure modeling using MODELLER. Curr Protoc Protein Sci 86:2.9.1-2.9.37
Zhang XC, Wang Z, Zhang X et al (2012) Evolutionary dynamics of protein domain architecture in plants. BMC Evol Biol 12:6. https://doi.org/10.1186/1471-2148-12-6
Zimmermann L, Stephens A, Nam SZ et al (2018) A completely reimplemented MPI bioinformatics toolkit with a new hhpred server at its core. J Mol Biol 2836(17):30587–30589. https://doi.org/10.1016/j.jmb.2017.12.007
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare that this article content has no conflict of interest.
Additional information
Handling editor: Arturo Becerra.
Supplementary Information
Below is the link to the electronic supplementary material.
239_2023_10109_MOESM1_ESM.txt
Supplementary file1 (TXT 404 kb). Supplementary file 1: The HEN1 homologous sequences from the plant species considered for this study
239_2023_10109_MOESM2_ESM.clans
Supplementary file2 (CLANS 1786 kb). Supplementary file 2: The constructed file of sequence-similarity-based grouping of the HEN1 protein family dataset by CLANS
239_2023_10109_MOESM3_ESM.txt
Supplementary file3 (TXT 287 kb). Supplementary file 3: The predicted functional domains using querying the protein sequence in the Hmmscan search tool (https://www.ebi.ac.uk/Tools/hmmer/search/hmmscan) against the Pfam database (http://pfam.xfam.org/)
239_2023_10109_MOESM4_ESM.pdf
Supplementary file4 (PDF 993 kb). Supplementary file 4: The unrooted phylogenetic tree of the HEN1 protein sequences was rooted with the outlier (outgroup Marchantia paleacea HEN1 protein sequence (KAG6553076.1)). Evolutionary analyses were conducted in MEGA11 and visualized by iTOL v5 online tool. In addition, graphical representation of the conserved motifs (B)/domains (C) variations and their architecture in the Plant HEN1 protein sequence and their evolutionary relationship within each HEN1 protein clade. The protein motifs were obtained using MEME suite (http://memesuite.org/) and the functional domains were predicted by querying the protein sequence in the Hmmscan search tool (https://www.ebi.ac.uk/Tools/hmmer/search/hmmscan) against the Pfam database (http://pfam.xfam.org/). Different motifs were represented by different colored boxes and the consensus sequences of the motifs are listed. The domains are marked with different shapes and colors
239_2023_10109_MOESM5_ESM.pdf
Supplementary file5 (PDF 190 kb). Supplementary file 5: Gene duplications were identified by searching for all branching points in the topology with at least one species that is present in both subtrees of the branching point. Evolutionary analyses were conducted in MEGA11
239_2023_10109_MOESM6_ESM.pdb
Supplementary file6 (PDB 566 kb). Supplementary File 6: The predicted PDB files for 20 HEN1selected proteins from the oldest extant land plant lineages to those present in modern HEN1 in angiosperms considered for this study
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hajieghrari, B., Niazi, A. Phylogenetic and Evolutionary Analysis of Plant Small RNA 2′-O-Methyltransferase (HEN1) Protein Family. J Mol Evol 91, 424–440 (2023). https://doi.org/10.1007/s00239-023-10109-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-023-10109-0