Abstract
In the past few years, our understanding of the RNA virosphere has changed dramatically due to the growth and spurt of metagenomics, exponentially increasing the number of RNA viral sequences, and providing a better understanding of their range of potential hosts. As of today, the only conserved protein among RNA viruses appears to be the monomeric RNA-dependent RNA polymerase. This enzyme belongs to the right-hand DNA-and RNA polymerases, which also includes reverse transcriptases and eukaryotic replicative DNA polymerases. The ubiquity of this protein in RNA viruses makes it a unique evolutionary marker and an appealing broad-spectrum antiviral target. In this work pairwise structural comparisons of viral RdRps and RTs were performed, including tertiary structures that have been obtained in the last few years. The resulting phylogenetic tree shows that the RdRps from (+)ss- and dsRNA viruses might have been recruited several times throughout the evolution of mobile genetic elements. RTs also display multiple evolutionary routes. We have identified a structural core comprising the entire palm, a large moiety of the fingers and the N-terminal helices of the thumb domain, comprising over 300 conserved residues, including two regions that we have named the “knuckles” and the “hypothenar eminence”. The conservation of an helix bundle in the region preceding the polymerase domain confirms that (−)ss and dsRNA Reoviruses’ polymerases share a recent ancestor. Finally, the inclusion of DNA polymerases into our structural analyses suggests that monomeric RNA-dependent polymerases might have diverged from B-family polymerases.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The development of metagenomics, metatranscriptomics and the powerful bioinformatic tools to explore these techniques has dramatically changed our understanding of the global RNA virosphere and has made clear that so far, we have merely characterized the tip of the iceberg (Shi et al. 2018a; Zhang et al. 2019; Edgar et al. 2022). The number and the diversity of sequenced RNA viruses has increased exponentially with each new published work (Shi et al. 2016, 2018a; Edgar et al. 2022). The diversity of hosts in which viruses have been discovered is also on the rise; interestingly, most of the new hosts are invertebrates (Shi et al. 2016) and basal vertebrate lineages such as fish and amphibians (Shi et al. 2018b; Parry et al. 2020), but this result may be the outcome of a sampling bias.
RNA viruses have also been at the public health spotlight for the last couple of years. The infamous COVID-19 pandemic is caused by the SARS-CoV-2, an RNA virus of the Coronaviridae family. As of February 2022, this emergent disease has left a toll of over 435 million cases and over 6 million deaths around the globe (https://coronavirus.jhu.edu/). Despite the plethora of information gathered, the available data suggest that the only conserved protein in all the sequenced RNA viruses is the monomeric RNA-dependent RNA polymerase (RdRp) (Shi et al. 2016; Zhang et al. 2019; Edgar et al. 2022). The ubiquity of the RdRp in RNA viruses makes it a molecular marker to infer deep evolutionary relationships between these viruses, retroviruses, and a set of DNA viruses endowed with homologous polymerases (Shi et al. 2016; Wolf et al. 2018). This has also led to consider it as an equally attractive target in the search for broad-spectrum antivirals (Cannalire et al. 2020; Jácome et al. 2020; Wang et al. 2021).
Monomeric RNA-dependent polymerases (Rdp) are part of a diverse group of enzymes that play an essential role in viral and cellular processes. These enzymes include the replicative RdRps of RNA viruses and retroviral reverse transcriptases (RT), the replicative enzymes of eukaryotic mobile genetic elements such as LTR- and non-TLR retrotransposons (Finnegan 2012), group II introns (Zhao and Pyle 2016) and diversity-generating retroelements (DGR) in prokaryotes (Novikova and Belfort 2017). In addition, cellular organisms from the three major domains also encode for different RTs performing a wide array of functions including defense systems in prokaryotes (Toro et al. 2019; González-Delgado et al. 2021), the extension of telomeres (Mitchell et al. 2010), and as part of the pre-mRNA-splicing factor 8 in the eukaryotic spliceosome (Galej et al. 2013).
All these polymerases are part of the Superfamily of DNA- and RNA polymerases, in which the fingers, palm and thumb functional subdomains can be identified (Steitz 1999). In the case of viral RdRps, the hand adopts a closed-hand shape due to the presence of structural elements, deemed the “fingertips”, that allow the interaction of the fingers and the thumb subdomains (Ferrer-Orta et al. 2006). The palm subdomain contains two universally conserved catalytic aspartic acids, which catalyze the synthesis of the 3′–5′ bond, and, perhaps not surprisingly, it is the most ancient and the most conserved one (Steitz 1999). Despite the primary structure variability in monomeric Rdps, their three-dimensional structures have revealed a high degree of structural conservation. Seven structural motifs (A–G) are conserved in monomeric Rdps. Each of these motifs include residues that participate in key steps, such as divalent metal ion-binding, discrimination and binding of the incoming nucleotide, and coordination of the leaving pyrophosphate group (Jácome et al. 2015). Bioinformatic methods have led to the recognition of an additional motif H, although as of today its function has not been elucidated (Cerny et al. 2014).
Except for several Nidovirales families, that include the Coronaviridae, RNA viruses are characterized by the lack of proofreading mechanisms during genome replication, which results in an extremely high mutation rate (10E−3–10E−5 mut/site/rep) in comparison to prokaryotes (Duffy 2018). Thus, the evolutionary relatedness between homologous proteins can rapidly become unidentifiable in terms of primary structure. Since the tertiary structure of proteins is more conserved in evolution, the use of tertiary structure-based phylogenies has become a valuable alternative when studying proteins for which the evolutionary relationships have become “hazy”, as is the case of viral monomeric Rdps (Cerni et al. 2014; Mönttinen et al. 2014, 2021; Jácome et al. 2015; Venkataraman et al. 2018; Peersen 2019).
Apart from their significance as a plausible broad-spectrum antiviral target, monomeric Rdps have also been the focus of research efforts on the origins and early evolution of life (Lazcano 1986; Lazcano et al. 1988). Despite being an open issue, it has been argued that prior to the evolutionary emergence of cellular DNA genomes, life probably went through a series of RNA-based stages, i.e. the RNA world, a very ancient and perhaps primordial life stage in which RNA partook in the transmission of genetic information as well as the catalytic capabilities of RNA-based entities, followed by an RNA–protein world, in which proteins gradually took over the catalytic repertoire and RNA served as the informational molecule (Gilbert 1986; Vásquez-Salazar and Lazcano 2018; Hernández-Morales et al. 2019). During these early stages, well before the emergence of the Last Universal Common Ancestor (LUCA), monomeric polymerases lacking absolute template and/or product specificity might have existed, and may have polymerized nucleic acids homologs that might have preceded RNA itself such as TNA (Chaput et al. 2003). As discussed below, minor changes in few key residues might have allowed for the emergence of more specific monomeric polymerases, such as replicative DNA-dependent DNA polymerases, monomeric DNA-dependent RNA polymerases, and RNA-dependent DNA polymerases, i.e. RTs. Prokaryotic group II introns RTs should have led to the emergence of eukaryotic proteins such as the telomerase and the spliceosomal prp8 (Doolittle 2014; Lambowitz and Belfort 2015; Novikova and Belfort 2017; González-Delgado et al. 2021). The analyses of biological information complementary to the phylogenetic hypotheses, hint to a more recent emergence of RdRp-dependent RNA viruses, perhaps at the evolution of eukaryotic cells (Campillo-Balderas et al. 2015). The versatility of polymerases is also present in the “double-psi β-barrel” family, in which replicative archaeal DNA polymerases, multi-subunit cellular RNA polymerases and eukaryotic RdRps share a homologous protein fold (Sauguet et al. 2016).
Since the publication of our previous work (Jácome et al. 2015), the structures of several RNA viral replicases and cellular RTs have been obtained, either by X-ray crystallography or by cryo-electron microscopy. Accordingly, we have decided to include these structures into our evolutionary analysis and provide here an updated version of the monomeric Rdp’s evolutionary tree. A careful analysis of the structures allowed us to identify additional universally conserved structural elements which we have named the “knuckles” and the “hypothenar eminence”, following the anatomically-based right-hand nomenclature used for this enzyme. Finally, we added cellular right-hand DdDps to a subset of the Rdps structure-based tree, yielding a scenario in which extant monomeric Rdps diverged from family B DdDps.
Material and Methods
Phylogenetic Trees
RNA-Dependent Monomeric Polymerases Structure Selection
A search in the Protein Data Bank (PDB) was performed using the terms “RNA-dependent RNA polymerase” (E.C. 2.7.7.48) and “reverse transcriptase” (E.C. 2.7.7.49). The structures that were released after the publication of our previous article (Jacome et al. 2015) and up to January 2022 were selected and added to the database. When more than one structure was available for the same polymerase, we selected those without ligands or bound substrates, those with the highest degree of completeness, or those with the highest structural resolution. The structures were edited so that only the polymerase domain was compared, therefore additional domains and accessory structural elements were not considered for the structural comparisons. The structures with poor resolution (> 4 Å) were discarded for further evolutionary analysis. Overall, fifty-four structures were included for the RdRPs and RTs’ structural comparisons. The entire list of the structures used in this section can be found as Supplementary Table 1.
DNA-Dependent DNA Polymerases Structure Selection
A search in the Protein Data Bank (PDB) was performed using the terms “DNA-dependent DNA polymerase” (E.C. 2.7.7.7). Structures from the three right-hand DNA polymerases families, i.e. family A, family B and family Y, were selected manually from the database. When more than one structure was available for the same polymerase, we selected those without ligands or bound substrates, those with the highest degree of completeness, or those with the highest resolution. As with the RdRps, structures with poor resolution (> 4A) were discarded for further evolutionary analyses. In the end, thirty-two DdDps (8 family A, 16 family B, 8 family Y) were added to a subset of 37 Rdp structures. The entire list of the structures used in this section can be found as Supplementary Table 2. The structures were edited so that only the polymerase domain was compared, therefore additional domains and accessory structural elements were not considered for the structural comparisons.
Structural Comparisons and Phylogenetic Trees Construction
The pairwise structural comparisons were performed with the web-based Secondary Structure Matching program (Krissinel and Henrick 2004), within the Protein Data Bank in Europe. From each pairwise comparison, we obtained the RMSD and the number of superimposed residues. In order to normalize the results, we calculated the Structural Alignment Score (Subbiah 1993) using the following formula: [(RMSD × 100)/number of superimposed residues]. Geometric distance matrices were built, one for the monomeric RNA-dependent polymerases and one that also included DdDps. These matrices were then processed with the FITCH algorithm included in the PHYLIP version 3.695 package to infer phylogenetic trees. The latter were visualized and edited with FigTree (http://tree.bio.ed.ac.uk/software/figtree/).
Structures’ Visualization, Analysis, and Rendering
All the structures were visualized, analyzed, and rendered with Chimera version 1.14 (Pettersen et al. 2004).
Results and Discussion
Updating the Rdps Phylogenetic Tree
In this evolutionary analysis, we have included 54 Rdps’ structures: 25 tertiary structures used in the previous publication (Jácome et al. 2015) and 29 new ones in this work. These recently added polymerase structures belong to: double-stranded RNA viruses (dsRNA): Cystoviridae (1), Picobirnaviridae (1), and Reoviridae (1); positive sense single-stranded RNA viruses [(+)ssRNA]: Picornaviridae (4), Caliciviridae (1), Coronaviridae (2), Permutotetraviridae (1), and Flaviviridae (3); negative sense single-stranded RNA viruses [(−)ssRNA]: Arenaviridae (1), Orthomyxoviridae (1), Phenuiviridae (1), Pneumoviridae (1), and Rhabdoviridae (2); single-stranded RNA viruses with RT (ssRNA-RT): Retroviridae (2) and Metaviridae (1). We have also added six cellular RTs: Tetrahymena thermophila- and Homo sapiens telomerase’s RT, three bacterial group II Introns, and Saccharomyces cerevisiae spliceosome’s Prp8.
Perhaps not surprisingly, the overall topology of the unrooted tree (Fig. 1) is in fact quite similar to those previously published (Cerni et al. 2014; Mönttinen et al. 2014, 2021; Jácome et al. 2015; Venkataraman et al. 2018). There are two branches that only include RdRps from (+)ssRNA viruses, one clustering families Picorna, Calici and Coronaviridae, and the other that encompasses the Flaviviridae family. Double-stranded RNA polymerases are found in several branches along the tree. The case of the Birna/Permutotetraviridae polymerases is interesting, since these polymerases are characterized by the presence of a circular permutation, in which the order of the strands that conform the palm subdomain is altered, instead of being 2-3-1-4, they are 1-2-3-4; the rest of the subdomains are highly conserved (Gorbalenya et al. 2002; Pan et al. 2005; Ferrero et al. 2015). Further in the tree there is a branch including the Cystoviridae, and another branch with dsRNA Reoviridae and (+)ssRNA Fiersviridae. Polymerases from (−)ssRNA viruses form one branch. Segmented (−)ssRNA viruses are, in turn, divided in Bunyavirales and Orthomyxoviridae branches, whereas the Mononegavirales are located in another clade, one branch corresponds to the Pneumoviridae, whereas the other branch groups all the Rhabdoviridae. The most divergent Rdps are the reverse transcriptases, which are all grouped in one large branch, except for the spliceosomal prp8, which is interspersed between the viral RdRps. This might be explained by the fact that only one of the active site aspartates can be identified, indicating that prp8 has lost its polymerization activity (Galej et al. 2013). Cellular RTs form two different clades, one includes group II intron maturases, whereas the other groups eukaryotic telomerases. The Retroviridae and the Metaviridae RTs form the farthest clade. It is interesting to highlight that dsRNA polymerases form several different clades, which suggests that viral RdRps lack substrate specificity and that viruses with dsRNA genomes might have hijacked the RdRp several times. Our structural phylogeny is in accordance with previous phylogenies (Xiong and Eickbush 1990; Nakamura et al. 1997; Gladyshev and Arkhipova 2011), in which viral RTs and LTR retrotransposons are closer to each other and cellular RTs stem from different branches closer to viral RdRps.
Unrooted dendogram based on the structural comparisons of monomeric RNA-dependent polymerases. The branches are colored as follows: (+)ssRNA viruses—blue; (−)ssRNA viruses—red; dsRNA viruses—green; cellular reverse transcriptases—purple; viral reverse transcriptases—gold. YFV yellow fever virus, JEV Japanese encephalitis virus, DENV dengue virus, ZIKV zika virus, CSFV classical swine fever virus, BVDV bovine viral diarrhea virus, HCV hepatitis C virus, RHDV rabbit hemorrhagic disease virus, FMDV foot-and-mouth disease virus, IPNV infectious pancreatic necrosis virus, IBDV infectious bursal disease virus, Ty3 Saccharomyces cerevisiae Ty3 retrotransposon, FIV feline immunodeficiency virus, HIV-1 human immunodeficiency virus 1, HIV-2 human immunodeficiency virus 2, MMLV Moloney murine leukemia virus, Prp8 spliceosomal protein prp8, VSV vesicular stomatitis virus, RSV respiratory syncytial virus, SFTSV severe fever with thrombocytopenia syndrome virus, BMCV Bombyx mori cypovirus (Color figure online)
The Structural Conservation of Monomeric RNA-Dependent Polymerases Extends Beyond Conserved Motifs A–H
The elevated mutation rate of RNA viruses, combined with the diversity in terms of complementary functional domains of Rdps, have hindered the attempts to analyze their evolution through sequence-based approaches. However, as will be discussed below, the actual conservation of Rdps is remarkable when their tertiary structures are compared.
The numerical analysis of the pairwise comparisons shows that the mean RMSD for all of them is 3.38 Å, and the mean number of aligned residues is 287. When only the RdRps are considered, the mean RMSD is 3.08 Å and 348.3 aligned residues; whereas when the RTs are compared, the mean RMSD is 2.89 Å and 201.7 superimposed residues.
Mönttinen et al. (2021) described a conserved core of 231 amino acids comparing only the RdRp structures, which is significantly smaller than the 348 residues core we report. This difference might be due to the different methodologies used. The mean number of superimposed residues when RdRps and RTs are compared reveals that the structural conservation must necessarily extend beyond the palm subdomain. Lang et al. (2013) previously identified additional conserved structures, which they named “homomorphs”, in the moieties preceding and following structural motifs A-G in the (+)ss and dsRNA polymerases available at the time of their publication. However, since that time, the number and the diversity of the polymerases’ three-dimensional structures has increased and now include milestones like the determination of various (−)ssRNA viral polymerases. We have therefore aimed to identify the additional conserved elements in all the tertiary structures of monomeric viral Rdps available by designating topologically equivalent structures, i.e. regions of the protein with the same secondary structure, located in a similar place within the amino acid primary structure, and with a similar orientation and connectivity within the tertiary structure. A thorough analysis of the RdRp and the RT structures suggests that these enzymes have a conserved core that extends beyond the structural motifs previously recognized (Fig. 2). The RMSD, the number of superimposed residues and the corresponding SAS from all the pairwise comparisons between RdRps and RTs can be found in Supplementary table 3.
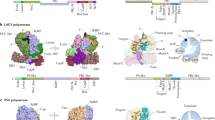
Extended conserved core in RNA-dependent RNA polymerases including the “knuckles” and “hypothenar eminence” structural motifs. a The Enterovirus D68 3Dpol (PDB 5XE0) is depicted here in “frontal” and “dorsal” views as a representative RdRp. Structural motifs A–F are colored orange, whereas the additional structural elements described in this work are colored blue. b From left to right, representative RdRp structures belonging to Baltimore groups III (dsRNA; PDB 6TZ0, Bombyx mori cypovirus; the comparison with 5XE0 yields the following results: RMSD—3.74; No. of superimposed residues—321; SAS—1.165), IV [(+)ss RNA; PDB 5XE0, Enterovirus D68] and V [(−)ssRNA; PDB 4WRT, Influenza B virus; the comparison with 5XE0 yields the following results: RMSD—3.75; No. of superimposed residues—300; SAS—1.250] are shown here. Structural motifs A–F are colored orange, the “knuckles” structural motif is colored purple, the “hypothenar eminence” structural motif is colored green, and the rest of the extended core is colored blue (Color figure online)
The Extended Conserved Core in RdRps
For the description of the extended conserved core, we will consider, as a three-dimensional reference, that the active site of the palm subdomain is facing upwards and motifs D and E are in the anterior part of the polymerase (Fig. 2a). In all the three-dimensional viral RdRp structures available, the extended conserved core includes several regions of the fingers, the palm domain, plus the N-terminal structural elements of the thumb subdomain. The average number of residues in the viral RdRps encompassing this extended conserved core is 424 residues and the most significant numbers are those of (−)ssRNA viruses, which are the ones with a higher number of additional structures.
The conserved core starts with a descending helix located above motif D followed by a small connector; next, there is a helix perpendicular to the palm subdomain that runs behind it which we have named the “knuckles” (Fig. 2b), followed by a connector whose average length is 9 residues going upwards lacking secondary structure, with the exception of (−)ssRNA viral polymerases, in which there are well-defined helical elements projecting outwards. The next moiety has been deemed the “pinky finger” and is part of the template entrance tunnel (Thompson et al. 2007). This region is quite variable in terms of structure, although there is a conserved structural element in (+)ss and some dsRNA viral polymerases which has been denominated motif G (Gorbalenya et al. 2002; Pan et al. 2005). In the case of (−)ssRNA polymerases, the characteristic signature of this motif (T/SX2G) (Gorbalenya et al. 2002) cannot be identified, and this region is longer, reaching over 120 amino acids in the Orthomyxoviridae PB1, consisting of long β-strands that extend above the core of the enzyme. This is followed by motif F, which partakes in the coordination of the incoming nucleotide’s phosphates via conserved positive residues (K or R). In the case of the Cystoviridae polymerases, following the first β-strand of motif F, there is an insertion of around 55 residues that projects towards the thumb subdomain in the exterior of the polymerase as part of the fingertips. After motif F, a “twisted” helix descends towards the palm subdomain followed by a connector facing the active site, and a second helix preceding motif A. Following the anatomical nomenclature, we named this ensemble the “hypothenar eminence” due to its location in the palm subdomain opposite the thumb subdomain (Fig. 2b). The only exceptions in terms of connectivity are the Birnaviridae and the Permutotetraviridae polymerases, in which the second helix connects with motif C and not with motif A due to a circular permutation (Pan et al. 2005; Ferrero et al. 2015). Right after motif A, perpendicular to the palm subdomain and above the “knuckles” helix, polymerases have a helix-turn-helix motif followed by a β-loop-β motif pointing towards the exterior of the protein. After the second strand there is a short connector, whose average length is 6 residues that leads to the highly conserved palm subdomain defined by structural motifs B to E. In (−)ssRNA polymerases, the region right after the first β-strand is bulkier due to an insertion of approximately 40 amino acids that points towards the surface of the protein. In all Rdps, the C-terminal moiety consists of the thumb subdomain, which interacts with the template and the primer chains. With very few exceptions, in DNA- and in RNA polymerases this subdomain is mostly helical and highly variable; nevertheless, in RdRps at least the first three structural elements are conserved and consist of three helices in the following direction: ascending-descending-ascending, and the second helix is located closer to the active site.
An Additional Conserved Structure in (−)ssRNA and Reoviridae Polymerases
The polymerases of (−)ssRNA viruses and dsRNA Reoviridae are considerably larger than those of ss(+) and the RTs, usually comprising several hundreds of residues and additional intricate structural elements surrounding the core subdomains. In our tree (Fig. 1), RdRps of these viruses form two distinct branches that are nevertheless close in terms of structural distance. In the region preceding the conserved core described above, and facing motifs D and E from the palm subdomain, these polymerases have a conserved helical bundle (Fig. 3). This conserved region had been previously recognized by Liang et al. (2015) in the polymerases of the Influenza A virus, the vesicular stomatitis virus and the Reovirus Lambda 3. Although the length of these helices is different in all the cases, and there are some additional structural elements in some of them, the connectivity and the relative location of the structures are the same (Fig. 3). In the case of (−)ssRNA Bunyavirales, Mononegavirales, and the dsRNA Reoviridae, this helical structure is located in the N-terminal half of a large polymerase protein; however, in the case of the Orthomyxoviridae, whose polymerases consist of three subunits (PA, PB1 and PB2), this fragment is formed by the C-terminal residues of the PA subunit and the N-terminal residues of the PB1 subunit. It must be underlined that in our tree, the branches including these viruses are consecutive. When a structural comparison is performed between the unedited Bunya, Mononega and Reoviridae polymerases, 200 additional residues can be superimposed, which should reduce the geometrical distance between them, and probably fuse the respective branches in the tree. This structural/evolutionary relatedness had been previously recognised by Wolf et al. (2018), albeit based on brittle sequence alignments, as will be discussed below, and by Mönttinen et al. (2021). Our work strengthens this hypothesis by showing the presence of conserved structural features in these viral families.
Helical bundle conserved in (−)ssRNA and dsRNA viral RdRps. The topologically equivalent helices are colored as follows (from the N-terminus to the C-terminus): orange, yellow, green, cyan, dark blue, purple, magenta. Representative RdRps shown here correspond to dsRNA virus: Simian rotavirus VP1 (PDB 2R7Q); segmented (−)ssRNA virus: Influenza B virus polymerase PDB 4WRT; non-segmented (−)ssRNA virus: respiratory syncytial virus L protein PDB 6PZK (Color figure online)
The Extended Conserved Core in Reverse Transcriptases
Previous sequence-based phylogenetic analysis divided RTs in two large groups, one encompassing viral RTs and LTR retrotransposons, and the other clustering group II introns and non-LTR retrotransposons (Xiong and Eickbush 1990). This divide is confirmed when the structures are visualized.
Cellular Reverse Transcriptases: Group II Introns Maturases, Spliceosomal Prp8 and Telomerase RT
Compared to RdRps, cellular RTs display a more diverse array of structures preceding Motif F, probably reflecting that each one of them participates in different cellular processes. The telomerase RTs have a helical RNA-binding domain (TRBD), which sits on top of the active site, and stabilizes the enzyme’s interactions with the nucleic acids (Mitchell et al. 2010; Gillis et al. 2010). On the other hand, the RTs of group II intron maturases and the spliceosomal Prp8 have a four-helical bundle above the fingers subdomain, which has been named N-Terminal Extension (NTE) or RT-0 (Zhao and Pyle 2016; Stamos et al. 2017).
Following motif F, cellular RTs present what we have termed the “hypothenar eminence” (Fig. 4b), called RT-2a in RTs (Stamos et al. 2017), which has a structure quite similar to viral RdRps, and consists of a descending twisted helix, a connector, and a helix prior to motif A (Fig. 4). Right after motif A, RTs also have the conserved structure consisting of a helix-turn-helix (RT-3a; Stamos et al. 2017) followed by a β hairpin, also named the Insertion in the Fingers Domain (IFD). The β hairpin in the R. intestinalis and E. faecalis group II intron maturases is in an inactive conformation, occupying the active site and not pointing towards the exterior of the protein (Zhao and Pyle 2016). Moreover, the thumb subdomain is absent in these group II intron RTs. The rest of the cellular RTs present the three-helical bundle in the N-terminus of the thumb subdomain (Fig. 4a); however, although the first two helices of the telomerase thumb have a similar location, their structural elements are not clearly defined (Fig. 4).
Extended conserved core in reverse transcriptases. a Tribolium castaneum telomerase reverse transcriptase (PDB 3KYL) is depicted on the left and human immunodeficiency virus 1 reverse transcriptase (PDB 4G1Q) is depicted on the right. The comparison between both structures yields the following results: RMSD—3.24; No. of superimposed residues—156; SAS—2.077. Structural motifs A–F are colored orange, whereas the additional structural elements described in this work are colored blue. b Tribolium castaneum telomerase reverse transcriptase (left) and human immunodeficiency virus 1 reverse transcriptase (right) showing the “hypothenar eminence” motif colored green (Color figure online)
Viral RTs
Viral RTs (Retroviridae and Metaviridae) are endowed with most of the extended conserved core of RdRps and cellular RTs, including structural motifs A–F (Figs. 2, 4). Many of the structural features of the extended conserved core preserve the same connectivity and location, but without clearly defined secondary structures. These regions comprise the helix-turn-helix following motif A and the hypothenar eminence or RT2a; in these segments, the helical elements are very small or substituted by connectors (Fig. 4b).
Additional features are more similar to the telomerase RT than to the other cellular RTs included in this work. The conserved helical bundle of NTE/RT-0 is substituted by a connector located above the active site and a helix located outside the protein above motifs D and E leading to motif F. Moreover, the first helix of the thumb subdomain is replaced by an ascending connector, in an analogous way to the telomerase.
Reconstructing the Global RNA Virosphere Evolution: Walking on Quicksand?
It is tempting to try to understand the evolution of all RNA viruses using the RdRp as an evolutionary marker (Wolf et al. 2018; Koonin et al. 2020; Edgar et al. 2022). However, the use of the RdRps primary structure is a double-edged sword. First of all, the available evidence suggests that RdRps have undergone polyphyletic recruitments by the ancestors of extant RNA viral groups. On the other hand, it allows to incorporate all the viral known sequences without the need for biochemical or molecular characterization, thereby, including metagenomic-derived putative viral sequences. However, as pointed by Holmes and Duchene (2019), sequence-based phylogenetic reconstructions have proven to have methodological limitations when studying deep evolutionary phenomena or highly diverging proteins, which is the case of RNA viral polymerases. Considering its intrinsic difficulties, it may be somewhat ambitious not only to posit evolutionary relations between highly divergent sequences, but also to propose a timeline for the evolutionary events that have led to the viral groups known today (Wolf et al. 2018; Koonin et al. 2020).
An additional caveat of some sequence-based evolutionary works (Wolf et al. 2018; Koonin et al. 2020) is that they have not included viral RTs in their analyses, although the latter are known to be homologous to the viral RdRps. Viral RTs could have been used as an outgroup to root the tree instead of the cellular counterparts. As noted by Zhao and Pyle (2016) and pointed out in this work, the structural similarity between viral RdRps and cellular RTs is quite remarkable; conversely, viral RTs are the farthest in terms of structural distance, whereas the eukaryotic telomerases are located somewhere in between (Fig. 1), sharing structural features with cellular and viral RTs.
Biochemical characterizations of RNA polymerases have shown that by mutating some key residues (Lyakhov et al. 1992; Sousa and Padilla 1995; Rai et al. 2017) or by substituting the divalent metal ion from Mg2+ to Mn2+, these enzymes can incorporate dNTPS and extend a DNA chain (Arnold et al. 1999; Hung et al. 2002). The same is true for DdDps, in which a few mutations in paramount residues (Gao et al. 1997; Xia et al. 2002; Cozens et al. 2012; Vaisman et al. 2012), or the substitution of Mn2+ instead of Mg2+ in the active site, favors the synthesis of RNA chains (Riccheti and Buc 1994). Extant RTs and replicative right-hand DdDps discriminate between ribonucleotides and deoxyribonucleotides by a single bulky residue located in the upper portion of motif A, which is called the “steric gate”: Glu in family A polymerases, and Tyr or Phe in family B DdDps and RTs (Gao et al. 1997; Astatke et al. 1998; Brown and Zuo 2011). This suggests that relatively minor changes in ancestral polymerases could have led to their adaptation to the different templates that may have appeared during cellular evolution. Recently, Peyambari et al. (2021) proved that the partitivirus polymerase (dsRNA plant-viruses that cause persistent infections) can carry out RNA-dependent RNA polymerization and reverse transcription. This dual function had not been previously reported in RNA viruses replicases, leading Peyambari et al. (2021) to propose that dsRNA viral polymerases with RdRp and RT activities might have been the first ones to emerge. Conversely, the identification in bacteria and archaea of numerous RTs associated with distinct functions (Toro and Nisa-Martínez 2014; Toro et al. 2019) has reinforced the hypothesis that prokaryotic group II introns were transferred to eukaryotes during the endosymbiosis processes via the ancestral mitochondria and chloroplasts, subsequently evolving into the spliceosome and the telomerase (Novikova and Belfort 2017; González-Delgado et al. 2021). The fact that our tree is unrooted does not allow us to assign a relative timeline for the evolutionary events observed. However, the works summarized here show that the right-hand polymerases’ substrate- and product specificities are far from absolute, and that minor changes allow them to use both RNA and DNA. Hence, it is likely that the earliest forms of the monomeric right-hand polymerases lacked absolute specificity, as has been suggested for other possible paramount ancient nucleic acid-associated enzymes such as the exonucleases (Zuckerkandl and Villet 1988; Dworkin et al. 2003; Cruz-González et al. 2021).
Structural Conservation in RNA-Dependent- and B-Family DNA Polymerases
We have included the DdDps that adopt a right-hand shape and are homologous to the viral RdRps and RTs, namely, family B, family A and family Y, and built a structure-based tree (Fig. 5). The RMSD, the number of superimposed residues and the corresponding SAS from all the pairwise comparisons between Rdps and DdDps can be found in Supplementary table 4. Based on the work by Mönttinen et al. (2016), we rooted the tree using the family Y polymerases as an outgroup. In their tree family B DdDps, family A DdDps, RdRps and RTs form one clade, whereas family Y DdDps group with enzymes such as nucleotide cyclases, Prim-pol domains, and Transposases IS200-like. In our tree (Fig. 5), family A pols, including the monomeric T7-phage monomeric DNA-dependent RNA polymerase are forming one branch. B-family DdDps are located on a different branch, from which monomeric Rdps stem.
Right-hand polymerases structure-based phylogenetic tree. The branches are colored as follows: cyan—family Y DNA-dependent DNA polymerases; orange—family A DNA-dependent DNA polymerases; purple—family B DNA-dependent DNA polymerases; gold—viral reverse transcriptases; magenta—cellular reverse transcriptases; green—ds RNA-dependent RNA polymerases; red—(−)ss RNA-dependent RNA polymerases; blue—(+)ss RNA-dependent RNA polymerases. DdDp DNA-dependent DNA polymerases, JEV Japanese encephalitis virus, DENV dengue virus, BVDV bovine viral diarrhea virus, HCV hepatitis C virus, RHDV rabbit hemorrhagic disease virus, FMDV foot-and-mouth disease virus, IPNV infectious pancreatic necrosis virus, IBDV infectious bursal disease virus, Ty3 Saccharomyces cerevisiae Ty3 retrotransposon, FIV feline immunodeficiency virus, HIV-1 human immunodeficiency virus 1, HIV-2 human immunodeficiency virus 2, MMLV Moloney murine leukemia virus, Prp8 spliceosomal protein prp8, VSV vesicular stomatitis virus, RSV respiratory syncytial virus, SFTSV severe fever with thrombocytopenia syndrome virus, BMCV Bombyx mori cypovirus (Color figure online)
A detailed analysis of the structures provides direct clues to understanding the relatedness between family B DdDps and monomeric Rdps (Fig. 6). On the one hand, despite the presence of additional structural and functional domains in many of these enzymes, the core of the polymerase domain is highly conserved including the entire palm subdomain. However, the subdomains’ order may provide insights on their evolutionary relationships. In family A polymerases, the thumb precedes the fingers and palm subdomains, whereas in family B and monomeric Rdps the sequence is as follows: fingers—motif A—fingers—motifs B–D—thumb. Moreover, in family B pols as well as in Rdps, the elements preceding motif A consist of a helix located in the fingers followed by a connector that descends towards the base of the palm, i.e. the hypothenar eminence (Fig. 6). As mentioned above, motif E in RNA polymerases, i.e. three small antiparallel beta-strands perpendicular to the main palm β-sheet, precedes three conserved helices of the thumb subdomain. Interestingly, B-family polymerases display a similar set of structural elements (Fig. 6). Three antiparallel β-strands follow the palm subdomain; however, in these polymerases, the strands are more extended, giving the appearance of two parallel β-sheets. These strands are followed by an “ascending” connecting element, which lacks any recognizable secondary structure in most of the available structures. Following this connector there are two antiparallel helices, the first one “descends” whereas the second “ascends”. Despite the differences in length of some of the B-family DdDps’ elements, the connectivity is practically the same compared to those of monomeric RNA polymerases.
Depiction of representative family B DNA-dependent DNA polymerases and RNA-dependent polymerases highlighting the topologically equivalent structural elements. a Thermococcus gorgonarius DNA-dependent DNA polymerase (PDB 1TGO; the comparison with 5XE0 yields the following results: RMSD—3.46; No. of superimposed residues—104; SAS—3.327); b Saccharomyces cerevisiae DNA-dependent DNA polymerase Delta (PDB 3IAY; the comparison with 5XE0 yields the following results: RMSD—4.09; No. of superimposed residues—129; SAS—3.170); c Enterovirus D68 RNA-dependent RNA polymerase (PDB 5XE0); d Human immunodeficiency virus 1 reverse transcriptase (PDB 4G1Q; the comparison with 5XE0 yields the following results: RMSD—5.5; No. of superimposed residues—161; SAS—3.416). In the 4 structures the colors are as follows: structural motifs A–D and F—orange; beta-strands preceding the thumb subdomain—cyan; “hypothenar eminence”—green; thumb subdomain N-terminal helices—magenta; fingers subdomain—dark blue; thumb subdomain—red (Color figure online)
B-family DdDps are distributed in all domains of life and in many DNA viruses. In eukaryotes, most archaea, and several double-stranded DNA viruses, they partake in genomic replication and repair, whereas in bacteria they participate in repair processes (Kazlauskas et al. 2020).
Conclusions
The significance of structure-based phylogenetic trees is increasingly becoming more evident, especially when studying deep evolutionary events such as the intricate evolution of right-hand DNA- and RNA polymerases. As shown in this work, despite their elevated mutation rates, viral RdRps and RTs as well as cellular RTs’ structures share an extensive, conserved core that can be easily identified when comparing three-dimensional structures. Unlike primary sequence-based methodologies, in which no more than a handful of conserved residues and small motifs can be identified, the conserved structural core comprises most of the fingers and the N-terminal moiety of the thumb subdomains, and the entire palm subdomain, which is the catalytic component involved in the formation of the 3′–5′ bonds. Our tree supports the idea that dsRNA as well as (+)ssRNA viral polymerases might have undergone polyphyletic recruitments during the evolutionary emergence of different viral groups, and that their template- and substrate specificities are far from absolute. Our results support the possibility that the Reoviridae polymerases and those of (−)ssRNA polymerases might share a recent ancestor. However, at the time being, the tree does not allow us to propose which came first. When monomeric viral Rdps and DdDps are compared, the tree indicates that the former diverged from the latter, which suggests that their emergence is more recent than previously thought, and that the biological entities encoding for them are not primordial.
The comparison of tertiary structures has proven quite successful for studying the evolutionary history of RNA viruses, whose extremely high mutation rates have erased the footprints within their primary structure. Analyses of tertiary structures have shown the wide array of evolutionary strategies exploited by RNA viruses such as gene duplications (Cisneros-Martínez et al. 2021) and hijacking events (Mönttinen et al. 2019; Cruz-González et al. 2021), underpinning the mosaic-nature of these biological entities’ genomes. The development and the advances of cryo-EM have been critical in the obtention of a more diverse spectrum of viral tertiary structures, and it is expected that this diversity will continue to increase, which might allow to answer some of the current evolutionary unknowns.
References
Arnold JJ, Ghosh SKB, Cameron CE (1999) Poliovirus RNA-dependent RNA polymerase (3Dpol): divalent cation modulation of primer, template and nucleotide selection. J Biol Chem 274:37060–37069
Astatke M, Ng K, Grindley NDF, Joyce CM (1997) A single side chain prevents Escherichia coli DNA polymerase I (Klenow fragment) from incorporating ribonucleotides. Proc Natl Acad Sci USA 95:3402–3407
Brown JA, Suo Z (2011) Unlocking the sugar “steric gate” of DNA polymerases. Biochemistry 50:1135–1142. https://doi.org/10.1021/bi101915z
Campillo-Balderas JA, Lazcano A, Becerra A (2015) Viral genome size distribution does not correlate with the antiquity of the host lineages. Front Ecol Evol 3:143. https://doi.org/10.3389/fevo.2015.00143
Cannalire R, Cerchia C, Beccari AR, Saverio Di Leva F, Summa V (2020) Targeting SARS-CoV-2 proteases and polymerase for COVID-19 treatment: state of the art and future opportunities. J Med Chem. https://doi.org/10.1021/acs.jmedchem.0c01140
Černý J, Černá Bolfíková B, Valdés JJ, Grubhoffer L, Růžek D (2014) Evolution of tertiary structure of viral RNA dependent polymerases. PLoS ONE 9(5):e96070
Chaput JC, Ichida JK, Szostak JW (2003) DNA polymerase-mediated DNA synthesis on a TNA template. J Am Chem Soc 125:856–857. https://doi.org/10.1021/ja028589k
Cisneros-Martínez AM, Becerra A, Lazcano A (2021) Ancient gene duplications in RNA viruses revealed by protein tertiary structure comparisons. Virus Evol 7(1):veab019. https://doi.org/10.1093/ve/veab019
Cozens C, Pinheiro VB, Vaisman A, Woodgate R, Holliger P (2012) A short adaptive path from DNA to RNA polymerases. Proc Natl Acad Sci USA 109:8067–8072. https://doi.org/10.1073/pnas.1120964109
Cruz-González A, Muñoz-Velasco I, Cottom-Salas W, Becerra A, Campillo-Balderas JA, Hernández-Morales R et al (2021) Structural analysis of viral ExoN domains reveals polyphyletic hijacking events. PLoS ONE 16(3):e0246981
Dolittle WF (2014) The trouble with (group II) introns. Proc Natl Acad Sci USA 111:6536–6537. https://doi.org/10.1073/pnas.1405174111
Duffy S (2018) Why are RNA virus mutation rates so damn high? PLoS Biol 16(8):e3000003. https://doi.org/10.1371/journal.pbio.3000003
Dworkin JP, Lazcano A, Miller SL (2003) The roads to and from the RNA world. J Theor Biol 222(1):127–134. https://doi.org/10.1016/S0022-5193(03)00020-1
Edgar RC, Taylor J, Lin V, Altman T, Barbera P, Meleshko D et al (2022) Petabase-scale sequence alignment catalyses viral discovery. Nature 602:142–147. https://doi.org/10.1038/s41586-021-04332-2
Ferrero D, Buxaderas M, Rodríguez JF, Verdaguer N (2015) The structure of the RNA-dependent RNA polymerase of a permutotetravirus suggests a link between primer-dependent and primer-independent polymerases. PLoS Pathog 11(12):e1005265. https://doi.org/10.1371/journal.ppat.1005265
Ferrer-Orta C, Arias A, Escarmís C, Verdaguer N (2006) A comparison of viral RNA-dependent RNA polymerases. Curr Opin Struct Biol 16:27–34. https://doi.org/10.1016/j.sbi.2005.12.002
Finnegan DJ (2012) Retrotransposons. Curr Biol 22(11):R432–R437
Galej WP, Oubridge C, Newman AJ, Nagai K (2013) Crystal structure of Prp8 reveals active site cavity of the spliceosome. Nature 493:638–643. https://doi.org/10.1038/nature11843
Gao G, Orlova M, Georgiadis MM, Hendrickson WA, Goff SP (1997) Conferring RNA polymerase activity to a DNA polymerase: a single residue in reverse transcriptase controls substrate selection. Proc Natl Acad Sci USA 94:407–411
Gilbert W (1986) Origin of life: the RNA world. Nature 319:618
Gillis AJ, Schuller AP, Skordalakes E (2008) Structure of the Tribolium castaneum telomerase catalytic subunit TERT. Nature 455:633–638. https://doi.org/10.1038/nature07283
Gladyshev EA, Arkhipova IR (2011) A widespread class of reverse transcriptase-related cellular genes. Proc Natl Acad Sci USA 108(51):20311–20316. https://doi.org/10.1073/pnas.1100266108
González-Delgado A, Rodríguez-Mestre M, Martínez-Abarca F, Toro N (2021) Prokaryotic reverse transcriptases: from retroelements to specialized defense systems. FEMS Microbiol Rev 25:1–19
Gorbalenya AE, Pringle FM, Zeddam JL, Luke BT, Cameron CE, Kalmakoff J et al (2002) The palm subdomain-based active site is internally permuted in viral RNA-dependent RNA polymerases of an ancient lineage. J Mol Biol 324:47–62
Hernández-Morales R, Becerra A, Lazcano A (2019) Alarmones as vestiges of a bygone RNA world. J Mol Evol 87:37–51. https://doi.org/10.1007/s00239-018-9883-3
Holmes EC, Duchene S (2019) Can sequence phylogenies safely infer the origin of the global virome? Mbio 10(2):e00289-19. https://doi.org/10.1128/mBio.00289-19
Hung M, Gibbs CS, Tsiang M (2002) Biochemical characterization of rhinovirus RNA-dependent RNA polymerase. Antiviral Res 56:99–114
Jácome R, Becerra A, Ponce de León S, Lazcano A (2015) Structural analysis of monomeric RNA-dependent polymerases: evolutionary and therapeutic implications. PLoS ONE 10(9):e0139001. https://doi.org/10.1371/journal.pone.0139001
Jácome R, Campillo-Balderas JA, Ponce de León S, Becerra A, Lazcano A (2020) Sofosbuvir as a potential alternative to treat the SARS-CoV-2 epidemic. Sci Rep 10:9294. https://doi.org/10.1038/s41598-020-66440-9
Kazlauskas D, Krupovic M, Guglielmini J, Forterre P, Venclovas C (2020) Diversity and evolution of B-family DNA polymerases. Nucl Acid Res 48:10142–10156. https://doi.org/10.1093/nar/gkaa760
Koonin EV, Dolja VV, Krupovic M, Varsani A, Wolf YI, Yutin N et al (2020) Global organization and proposed megataxonomy of the virus world. Microbiol Mol Biol Rev 84:e00061-19. https://doi.org/10.1128/MMBR.00061-19
Krissinel E, Henrick K (2004) Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr 60(Pt 12 Pt 1):2256–2268. https://doi.org/10.1107/S0907444904026460
Lambowitz AM, Belfort M (2015) Mobile bacterial group II introns at the crux of eukaryotic evolution. Microbiol Spectr. https://doi.org/10.1128/microbiolspec.MDNA3-0050-2014
Lang DM, Zemla AT, Zhou CLE (2013) Highly similar structural frames link the template tunnel and NTP entry tunnel to the exterior surface in RNA-dependent RNA polymerases. Nucl Acid Res 41:1464–1482. https://doi.org/10.1093/nar/gks1251
Lazcano A (1986) Prebiotic evolution and the origin of cells. Treballs De La Societat Catalana De Biología 39:73–103
Lazcano A, Fastag J, Gariglio P, Ramirez C, Oró J (1988) On the early evolution of RNA polymerase. J Mol Evol 27:365–376. https://doi.org/10.1007/BF02101199
Liang B, Li Z, Jenni S, Rahmeh AA, Morin BM, Grant T et al (2015) Structure of the L protein of vesicular stomatitis virus from electron cryomicroscopy. Cell 162:314–327. https://doi.org/10.1016/j.cell.2015.06.018
Lyakhov DL, Rechinsky VO, Tunitskaya VL, Chernov BK, Kochetkov SN (1992) Site-directed mutagenesis of bacteriophage T7 DNA-dependent RNA polymerase. J Cell Biochem 50(S16E):166
Mitchell M, Gillis A, Futahashi M, Fujiwara H, Skordalakes E (2010) Structural basis for telomerase catalytic subunit TERT binding to RNA template and telomeric DNA. Nat Struct Mol Biol 17:513–518. https://doi.org/10.1038/nsmb.3224
Mönttinen HAM, Ravantti JJ, Stuart DI, Poranen MM (2014) Automated structural comparisons clarify the phylogeny of the right-hand-shape polymerases. Mol Biol Evol 31:2741–2752. https://doi.org/10.1093/molbev/msu219
Mönttinen HAM, Ravantti JJ, Poranen MM (2016) Common structural core of three-dozen residues reveals intersuperfamily relationships. Mol Biol Evol 33:1697–1710. https://doi.org/10.1093/molbev/msw047
Mönttinen HAM, Ravantti JJ, Poranen MM (2019) Structural comparison strengthens the higher-order classification of proteases related to chymotrypsin. PLoS One 14(5):e0216659. https://doi.org/10.1371/journal.pone.0216659
Mönttinen HAM, Ravantti JJ, Poranen MM (2021) Structure unveils relationships between RNA virus polymerases. Viruses 13:313. https://doi.org/10.3390/v13020313
Nakamura TM, Morin GB, Chapman KB, Weinrich SL, Andrews WH, Lingner J et al (1997) Telomerase catalytic subunit homologs from fission yeast and human. Science 277:955–959
Novikova O, Belfort M (2017) Mobile group II introns as ancestral eukaryotic elements. Trends Genet 33(11):773–783. https://doi.org/10.1016/j.tig.2017.07.009
Pan J, Vakharia VN, Tao YJ (2007) The structure of a birnavirus polymerase reveals a distinct active site topology. Proc Natl Acad Sci USA 166:1–6
Parry R, Wille M, Turnbull OMH, Geoghegan JL, Holmes EC (2020) Divergent influenza-like viruses of amphibians and fish support an ancient evolutionary history. Viruses 12:1042. https://doi.org/10.3390/v12091042
Peersen OB (2019) A comprehensive superposition of viral polymerase structures. Viruses 11:745. https://doi.org/10.3390/v11080745
Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem 25(13):1605–1612
Peyambari M, Guan S, Roossinck MJ (2021) RdRp or RT, that is the question. Mol Biol Evol 38:5082–5091. https://doi.org/10.1093/molbev/msab235
Rai DK, Diaz-San Segundo F, Campagnola G, Keith A, Schafer EA, Kloc A et al (2017) Attenuation of foot-and-mouth disease virus by engineered viral polymerase fidelity. J Virol 91:e00081-17. https://doi.org/10.1128/JVI.00081-17
Riccheti M, Buc H (1993) E. coli DNA polymerase I as a reverse transcriptase. EMBO J 12:387–396
Sauguet L, Raia P, Henneke G, Delarue M (2016) Shared active site architecture between archaeal PolD and multi-subunit RNA polymerases revealed by X-ray crystallography. Nature Commun 7:12227. https://doi.org/10.1038/ncomms12227
Shi M, Lin XD, Tian JH, Chen LJ, Chen X, Li CX et al (2016) Redefining the invertebrate RNA virosphere. Nature 540:539–543. https://doi.org/10.1038/nature20167
Shi M, Zhang YZ, Holmes EC (2018a) Meta-transcriptomics and the evolutionary biology of RNA viruses. Virus Res 243:83–90. https://doi.org/10.1016/j.virusres.2017.10.016
Shi M, Lin XD, Chen X, Tian JH, Chen LJ, Li K et al (2018b) The evolutionary history of vertebrate RNA viruses. Nature 556:197–202. https://doi.org/10.1038/s41586-018-0012-7
Sousa R, Padilla R (1995) A mutant T7 RNA polymerase as a DNA polymerase. EMBO J 14:4609–4621
Stamos JL, Lentzsch AM, Lambowitz AM (2017) Structure of a thermostable group II intron reverse transcriptase with template-primer and its functional and evolutionary implications. Mol Cell 68:926-939.e4. https://doi.org/10.1016/j.molcel.2017.10.024
Steitz TA (1999) DNA polymerases: structural diversity and common mechanisms. J Biol Chem 274:17395–17398
Subbiah S, Laurents DV, Levitt M (1993) Structural similarity of DNA-binding domains of bacteriophage repressors and the globin core. Curr Biol 3:141–148
Thompson AA, Albertini RA, Peersen OB (2007) Stabilization of poliovirus polymerase by NTP binding and fingers-thumb interactions. J Mol Biol 366:1459–1474. https://doi.org/10.1016/j.jmb.2006.11.070
Toro N, Nisa-Martínez R (2014) Comprehensive phylogenetic analysis of bacterial reverse transcriptases. PLoS ONE 9(11):e114083. https://doi.org/10.1371/journal.pone.0114083
Toro N, Martínez-Abarca F, Rodríguez-Mestre M, González-Delgado A (2019) Multiple origins of reverse transcriptases linked to CRISPR-Cas systems. RNA Biol 16(10):1486–1493. https://doi.org/10.1080/15476286.2019.1639310
Vaisman A, Kuban W, McDonald JP, Karata K, Yang W, Goodman MF, Woodgate R (2012) Critical amino acids in Escherichia coli UmuC responsible for sugar discrimination and base-substitution fidelity. Nucl Acid Res 40:6144–6157. https://doi.org/10.1093/nar/gks233
Vázquez-Salazar A, Lazcano A (2018) Early life: embracing the RNA world. Curr Biol 28(5):R220–R222. https://doi.org/10.1016/j.cub.2018.01.055
Venkataraman S, Prasad BVLS, Selvarajan R (2018) RNA dependent RNA polymerases: insights from structure. Funct Evol Virus 10:76. https://doi.org/10.3390/v10020076
Wang Y, Anirudhan V, Du R, Cui Q, Rong L (2021) RNA-dependent RNA polymerase of SARS-CoV-2 as a therapeutic target. J Med Virol 93:300–310. https://doi.org/10.1002/jmv.26264
Wolf YI, Kazlauskas D, Iranzo J, Lucia-Sanz A, Kuhn JH, Krupovic M, Dolja VV, Koonin EV (2018) Origins and evolution of the global RNA virome. Mbio 9:e02329-18. https://doi.org/10.1128/mBio.02329-18
Xia G, Chen L, Sera T, Fa M, Schultz PG, Romesberg FE (2002) Directed evolution of novel polymerase activities: mutation of a DNA polymerase into an efficient RNA polymerase. Proc Natl Acad Sci USA 99:6597–6602
Xiong Y, Eickbush TH (1990) Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J 9(10):3353–3362
Zhang YZ, Chen YM, Wang W, Qin XC, Holmes EC (2019) Expanding the RNA virosphere by unbiased metagenomics. Ann Rev Virol 6:119–139. https://doi.org/10.1146/annurev-virology-092818-015851
Zhao C, Pyle AM (2016) Crystal structures of a group II intron maturase reveal a missing link in spliceosome evolution. Nat Struct Mol Biol 23:558–565. https://doi.org/10.1038/nsmb.3224
Zuckerkandl E, Villet R (1988) Generation of high specificity of effect through low-specificity binding of proteins to DNA. FEBS Lett 231:298
Acknowledgements
We are indebted to Dr. Ricardo Hernández Morales for his contributions to this work. Support from the Dirección General de Personal Académico DGAPA-PAPIIT (IN214421) and DGAPA-PAPIME (PE204921) of the Universidad Nacional Autónoma de México is gratefully acknowledged.
Funding
This work was supported by DGAPA-PAPIIT (IN214421) and DGAPA-PAPIME (PE204921) of Universidad Nacional Autónoma de México.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Additional information
Handling editor: Aaron Goldman.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Jácome, R., Campillo-Balderas, J.A., Becerra, A. et al. Structural Analysis of Monomeric RNA-Dependent Polymerases Revisited. J Mol Evol 90, 283–295 (2022). https://doi.org/10.1007/s00239-022-10059-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-022-10059-z