Abstract
In the biosynthesis of terpenoids, the ample catalytic versatility of terpene synthases (TPS) allows the formation of thousands of different molecules. A steadily increasing number of sequenced plant genomes invariably show that the TPS gene family is medium to large in size, comprising from 30 to 100 functional members. In conifers, TPSs belonging to the gymnosperm-specific TPS-d subfamily produce a complex mixture of mono-, sesqui-, and diterpenoid specialized metabolites, which are found in volatile emissions and oleoresin secretions. Such substances are involved in the defence against pathogens and herbivores and can help to protect against abiotic stress. Oleoresin terpenoids can be also profitably used in a number of different fields, from traditional and modern medicine to fine chemicals, fragrances, and flavours, and, in the last years, in biorefinery too. In the present work, after summarizing the current views on the biosynthesis and biological functions of terpenoids, recent advances on the evolution and functional diversification of plant TPSs are reviewed, with a focus on gymnosperms. In such context, an extensive characterization and phylogeny of all the known TPSs from different Pinus species is reported, which, for such genus, can be seen as the first effort to explore the evolutionary history of the large family of TPS genes involved in specialized metabolism. Finally, an approach is described in which the phylogeny of TPSs in Pinus spp. has been exploited to isolate for the first time mono-TPS sequences from Pinus nigra subsp. laricio, an ecologically important endemic pine in the Mediterranean area.

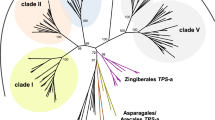
Adapted from Tholl et al. (2015)





Similar content being viewed by others
Data Availability
The cDNA and genomic sequences of the monoterpene synthases (MTPSs) from Pinus nigra subsp. laricio will appear in the GenBank database with the following accession numbers: MN088807 (Pnl_MBOS_1.1), MN088808 (Pnl_MTPS_1.2), MN088809 (Pnl_MTPS_1.5), MN088810 (Pnl_MTPS_1.7), MN088811 (Pnl_MTPS_1.4) and MN088812 (Pnl_MTPS_1.3).
References
Abbas F, Ke Y, Yu R, Yue Y, Amanullah S, Jahangir MM, Fan Y (2017) Volatile terpenoids: multiple functions, biosynthesis, modulation and manipulation by genetic engineering. Planta 246:803–816. https://doi.org/10.1007/s00425-017-2749-x
Aharoni A, Giri AP, Verstappen FWA, Bertea CM, Sevenier R (2004) Gain and loss of fruit flavor compounds produced by wild and cultivated strawberry species. Plant Cell 16:3110–3131. https://doi.org/10.1105/tpc.104.023895.native
Akhtar TA, Matsuba Y, Schauvinhold I, Yu G, Lees HA, Klein SE, Pichersky E (2013) The tomato cis-prenyltransferase gene family. Plant J 73:640–652. https://doi.org/10.1111/tpj.12063
Akiyama K, Matsuzaki K, Hayashi H (2005) Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 435:824. https://doi.org/10.1038/nature03608
Aubourg S, Lecharny A, Bohlmann J (2002) Genomic analysis of the terpenoid synthase (AtTPS) gene family of Arabidopsis thaliana. Mol Genet Genomics 267:730–745. https://doi.org/10.1007/s00438-002-0709-y
Baier M, Hartung W (1988) Movement of abscisic acid across the plasmalemma and the tonoplast of guard cells of Valerianella locusta. Plant Biol 101:332–337. https://doi.org/10.1111/j.1438-8677.1988.tb00053.x
Baldwin IT, Halitschke R, Paschold A, Von Dahl CC, Preston CA (2006) Volatile signaling in plant–plant interactions: “talking trees” in the genomics era. Science 311:812–815. https://doi.org/10.1126/science.1118446
Behnke K, Wildt R, Kleist E, Uerlings R (2009) RNAi-mediated suppression of isoprene biosynthesis in hybrid poplar impacts ozone tolerance. Tree Physiol 29:725–736. https://doi.org/10.1093/treephys/tpp009
Bick JA, Lange BM (2003) Metabolic cross talk between cytosolic and plastidial pathways of isoprenoid biosynthesis: unidirectional transport of intermediates across the chloroplast envelope membrane. Arch Biochem Biophys 415:146–154. https://doi.org/10.1016/S0003-9861(03)00233-9
Birol I, Raymond A, Jackman SD, Pleasance S, Coope R, Taylor GA et al (2013) Genome analysis assembling the 20 Gb white spruce (Picea glauca) genome from whole-genome shotgun sequencing data. Bioinformatics 29:1492–1497. https://doi.org/10.1093/bioinformatics/btt178
Bleeker PM, Diergaarde PJ, Ament K, Schütz S, Johne B, Dijkink J et al (2011) Tomato-produced 7-epizingiberene and R-curcumene act as repellents to whiteflies. Phytochemistry 72:68–73. https://doi.org/10.1016/j.phytochem.2010.10.014
Bohlmann J, Keeling CI (2008) Terpenoid biomaterials. Plant J 54:656–669. https://doi.org/10.1111/j.1365-313X.2008.03449.x
Bohlmann J, Steele CL, Croteau R (1997) Monoterpene synthases from grand fir (Abies grandis). cDNA isolation, characterization, and functional expression of myrcene synthase, (−)-(4S)-limonene synthase, and (−)-(1S,5S)-pinene synthase. J Biol Chem 272:21784–21792. https://doi.org/10.1074/jbc.272.35.21784
Bohlmann J, Meyer-Gauen G, Croteau R (1998) Plant terpenoid synthases: molecular biology and phylogenetic analysis. Proc Natl Acad Sci USA 95:4126–4133. https://doi.org/10.1073/pnas.95.8.4126
Bohlmann J, Phillips M, Ramachandiran V, Katoh S, Croteau R (1999) cDNA cloning, characterization, and functional expression of four new monoterpene synthase members of the Tpsd gene family from grand fir (Abies grandis). Arch Biochem Biophys 368:232–243. https://doi.org/10.1006/abbi.1999.1332
Büchel K, Malskies S, Mayer M, Fenning TM, Gershenzon J, Hilker M, Meiners T (2011) How plants give early herbivore alert: volatile terpenoids attract parasitoids to egg-infested elms. Basic Appl Ecol 12:403–412. https://doi.org/10.1016/j.baae.2011.06.002
Cao R, Zhang Y, Mann FM, Huang C, Mukkamala D, Hudock MP et al (2010) Diterpene cyclases and the nature of the isoprene fold. Proteins 78:2417–2432. https://doi.org/10.1002/prot.22751
Celedon JM, Bohlmann J (2019) Oleoresin defenses in conifers: chemical diversity, terpene synthases, limitations of oleoresin defense under climate change. N Phytol 224:1444–1463. https://doi.org/10.1111/nph.15984
Chen F, Pichersky E, Ro D, Petri J, Gershenzon J, Tholl D (2004) characterization of a root-specific Arabidopsis terpene synthase responsible for the formation of the volatile monoterpene 1, 8-cineole. Plant Physiol 135:1956–1966. https://doi.org/10.1104/pp.104.044388.1956
Chen F, Tholl D, Bohlmann J, Pichersky E (2011) The family of terpene synthases in plants: a mid-size family of genes for specialized metabolism that is highly diversified throughout the kingdom. Plant J 66:212–229. https://doi.org/10.1111/j.1365-313X.2011.04520.x
Cherian S, Ryu SB, Cornish K (2019) Natural rubber biosynthesis in plants, the rubber transferase complex, and metabolic engineering progress and prospects. Plant Biotechnol J 1–21: https://doi.org/10.1111/pbi.13181
Christianson DW (2017) Structural and chemical biology of terpenoid cyclases. Chem Rev 117:11570–11648. https://doi.org/10.1021/acs.chemrev.7b00287
Cordoba E, Salmi M, Leo P (2009) Unravelling the regulatory mechanisms that modulate the MEP pathway in higher plants. J Exp Bot 60:2933–2943. https://doi.org/10.1093/jxb/erp190
Dellas N, Thomas ST, Manning G, Noel JP (2013) Discovery of a metabolic alternative to the classical mevalonate pathway. Elife 2:e00672. https://doi.org/10.7554/eLife.00672
Dong L, Jongedijk E, Bouwmeester H, Krol A (2016) Monoterpene biosynthesis potential of plant subcellular compartments. N Phytol 209:679–690. https://doi.org/10.1111/nph.13629
Dudareva N, Flower C, Cseke L, Blanc VM, Pichersky E (1996) Evolution of floral scent in Clarkia: novel patterns of S-linalool synthase gene expression in the C. breweri flower. Plant Cell 8:1137–1148. https://doi.org/10.1105/tpc.8.7.1137
Dudareva N, Andersson S, Orlova I, Gatto N, Reichelt M, Rhodes D et al (2005) The nonmevalonate pathway supports both monoterpene and sesquiterpene formation in snapdragon flowers. Proc Natl Acad Sci USA 102:933–938. https://doi.org/10.1073/pnas.0407360102
Dudareva N, Klempien A, Muhlemann K, Kaplan I (2013) Tansley review biosynthesis, function and metabolic engineering of plant volatile organic compounds. N Phytol 198:16–32. https://doi.org/10.1111/nph.12145
Falara V, Akhtar TA, Nguyen TTH, Spyropoulou EA, Bleeker PM, Schauvinhold I et al (2011) The tomato terpene synthase gene family. Plant Physiol 157:770–789. https://doi.org/10.1104/pp.111.179648
Flügge U, Gao W (2005) Transport of isoprenoid intermediates across chloroplast envelope membranes. Plant Biol 7:91–97. https://doi.org/10.1055/s-2004-830446
Gao Y, Honzatko RB, Peters RJ (2012) Terpenoid synthase structures: a so far incomplete view of complex catalysis. Nat Prod Rep 29:1153–1175. https://doi.org/10.1039/C2NP20059G
Gennadios HA, Gonzalez V, Costanzo L, Li A, Yu F, Allemann RK et al (2009) Crystal structure of (+)-δ-cadinene synthase from Gossypium arboreum and evolutionary divergence of metal binding motifs for catalysis. Biochemistry 48:6175–6183. https://doi.org/10.1021/bi900483b
Ghassemian M, Lutes J, Tepperman JM, Chang HS, Zhu T, Wang X, Quail PH, Lange BM (2006) Integrative analysis of transcript and metabolite profiling data sets to evaluate the regulation of biochemical pathways during photomorphogenesis. Arch Biochem Biophys 448:45–59. https://doi.org/10.1016/j.abb.2005.11.020
Gols R (2014) Direct and indirect chemical defences against insects in a multitrophic framework. Plant Cell Environ 37:1741–1752. https://doi.org/10.1111/pce.12318
Gray DW, Breneman SR, Topper LA, Sharkey TD (2011) Biochemical characterization and homology modeling of methylbutenol synthase and implications for understanding hemiterpene synthase evolution in plants. J Biol Chem 286:20582–20590. https://doi.org/10.1074/jbc.M111.237438
Guirimand G, Guihur A, Phillips MA, Oudin A, Glevarec G, Melin C, Papon N, Clastre M, St-Pierre B, Rodríguez-Concepción M, Burlat V, Courdavault V (2012) A single gene encodes isopentenyl diphosphate isomerase isoforms targeted to plastids, mitochondria and peroxisomes in Catharanthus roseus. Plant Mol Biol 79:443–459. https://doi.org/10.1007/s11103-012-9923-0
Gutensohn M, Orlova I, Nguyen TTH, Davidovich-Rikanati R, Ferruzzi MG, Sitrit Y et al (2013) Cytosolic monoterpene biosynthesis is supported by plastid-generated geranyl diphosphate substrate in transgenic tomato fruits. Plant J 75:351–363. https://doi.org/10.1111/tpj.12212
Hall DE, Robert JA, Keeling CI, Domanski D, Quesada AL, Jancsik S et al (2011) An integrated genomic, proteomic and biochemical analysis of (+)-3-carene biosynthesis in Sitka spruce (Picea sitchensis) genotypes that are resistant or susceptible to white pine weevil. Plant J 65:936–948. https://doi.org/10.1111/j.1365-313X.2010.04478.x
Hall DE, Oa DSW, Zerbe P, Jancsik S, Quesada AL, Dullat H, Madilao LL (2013a) Evolution of conifer diterpene synthases: diterpene resin acid biosynthesis in lodgepole pine and jack pine involves monofunctional and bifunctional diterpene synthases. Plant Physiol 161:600–616. https://doi.org/10.1104/pp.112.208546
Hall DE, Yuen MM, Jancsik S, Quesada AL, Dullat HK, Li M et al (2013b) Transcriptome resources and functional characterization of monoterpene synthases for two host species of the mountain pine beetle, lodgepole pine (Pinus contorta) and jack pine (Pinus banksiana). BMC Plant Biol 13:80. https://doi.org/10.1186/1471-2229-13-80
Hamberger B, Hall D, Yuen M, Oddy C, Hamberger B, Keeling CI et al (2009) Targeted isolation, sequence assembly and characterization of two white spruce (Picea glauca) BAC clones for terpenoid synthase and cytochrome P450 genes involved in conifer defence reveal insights into a conifer genome. BMC Plant Biol 9:106. https://doi.org/10.1186/1471-2229-9-106
Hampel D, Mosandl A, Wust M (2005) Biosynthesis of mono- and sesquiterpenes in carrot roots and leaves (Daucus carota L.): metabolic cross talk of cytosolic mevalonate and plastidial methylerythritol phosphate pathways. Phytochemistry 66:305–311. https://doi.org/10.1016/j.phytochem.2004.12.010
Hayashi K, Kawaide H, Notomi M, Sakigi Y, Matsuo A, Nozaki H (2006) Identification and functional analysis of bifunctional ent-kaurene synthase from the moss Physcomitrella patens. FEBS Lett 580:6175–6181. https://doi.org/10.1016/j.febslet.2006.10.018
Hayashi K, Horie K, Hiwatashi Y, Kawaide H, Yamaguchi S (2010) Endogenous diterpenes derived from ent-kaurene, a common gibberellin precursor, regulate protonema differentiation of the moss Physcomitrella patens. Plant Physiol 153:1085–1097. https://doi.org/10.1104/pp.110.157909
Heil M (2014) Herbivore-induced plant volatiles: targets, perception and unanswered questions. N Phytol 204:297–306. https://doi.org/10.1111/nph.12977
Hemmerlin A (2013) Post-translational events and modifications regulating plant enzymes involved in isoprenoid precursor biosynthesis. Plant Sci 203:41–54. https://doi.org/10.1016/j.plantsci.2012.12.008
Hemmerlin A, Hoeffler JF, Meyer O, Tritsch D, Kagan IA, Grosdemange-Billiard C, Rohmer M, Bach TJ (2003) Cross-talk between the cytosolic mevalonate and the plastidial methylerythritol phosphate pathways in tobacco bright yellow-2 cells. J Biol Chem 278:26666–26676. https://doi.org/10.1074/jbc.M302526200
Henry LK, Gutensohn M, Thomas ST, Noel JP, Dudareva N (2015) Orthologs of the archaeal isopentenyl phosphate kinase regulate terpenoid production in plants. Proc Natl Acad Sci USA 112:10050–10055. https://doi.org/10.1073/pnas.1504798112
Henry LK, Thomas ST, Widhalm JR, Lynch JH, Davis TC, Kessler SA, Bohlmann J, Noel JP, Dudareva N (2018) Contribution of isopentenyl phosphate to plant terpenoid metabolism. Nat Plants 4:721–729. https://doi.org/10.1038/s41477-018-0220-z
Herde M, Fode B, Boland W, Gershenzon J, Ga K, Gatz C, Tholl D (2008) Identification and regulation of TPS04/GES, an Arabidopsis geranyl linalool synthase catalyzing the first step in the formation of the insect-induced volatile C16-homoterpene TMTT. Plant Cell 20:1152–1168. https://doi.org/10.1105/tpc.106.049478
Hillwig ML, Xu M, Toyomasu T, Tiernan MS, Wei G, Cui G et al (2011) Domain loss has independently occurred multiple times in plant terpene synthase evolution. Plant J 68:1051–1060. https://doi.org/10.1111/j.1365-313X.2011.04756.x
Hu XG, Liu H, Jin YQ, Sun YQ, Li Y, Zhao W et al (2016) De novo transcriptome assembly and characterization for the widespread and stress-tolerant conifer Platycladus orientalis. PLoS ONE 11:1–19. https://doi.org/10.1371/journal.pone.0148985
Huang M, Abel C, Sohrabi R, Petri J, Haupt I, Cosimano J et al (2010) Variation of herbivore-induced volatile terpenes among Arabidopsis ecotypes depends on allelic differences and subcellular targeting of two terpene. Plant Physiol 153:1293–1310. https://doi.org/10.1104/pp.110.154864
Huang M, Sanchez-Moreiras AM, Abel C, Sohrabi R, Lee S, Gershenzon J, Tholl D (2012) The major volatile organic compound emitted from Arabidopsis thaliana flowers, the sesquiterpene (E)-β-caryophyllene, is a defense against a bacterial pathogen. N Phytol 193:997–1008. https://doi.org/10.1111/j.1469-8137.2011.04001.x
Huber DP, Philippe RN, Godard KA, Sturrock RN, Bohlmann J (2005) Characterization of four terpene synthase cDNAs from methyl jasmonate-induced Douglas-fir, Pseudotsuga menziesii. Phytochemistry 66:1427–1439. https://doi.org/10.1016/j.phytochem.2005.04.030
Hyatt DC, Croteau R (2005) Mutational analysis of a monoterpene synthase reaction: altered catalysis through directed mutagenesis of (−)-pinene synthase from Abies grandis. Arch Biochem Biophys 439:222–233. https://doi.org/10.1016/j.abb.2005.05.017
Hyatt DC, Youn B, Zhao Y, Santhamma B, Coates RM, Croteau RB (2007) Structure of limonene synthase, a simple model for terpenoid cyclase catalysis. Proc Natl Acad Sci USA 104:5360–5365. https://doi.org/10.1073/pnas.0700915104
Irmisch S, Chen F, Gershenzon J, Jiang Y, Köllner TG (2014) Terpene synthases and their contribution to herbivore-induced volatile emission in western balsam poplar (Populus trichocarpa). BMC Plant Biol 14:1–16. https://doi.org/10.1186/s12870-014-0270-y
Jia Q, Köllner TG, Gershenzon J, Chen F (2018) MTPSLs: new terpene synthases in non seed plants. Trends Plant Sci 23:121–128. https://doi.org/10.1016/j.tplants.2017.09.014
Kai G, Xu H, Zhou C, Liao P, Xiao J, Luo X, You L, Zhang L (2011) Metabolic engineering Tanshinone biosynthetic pathway in Salvia miltiorrhiza hairy root cultures. Metab Eng 13:319–327. https://doi.org/10.1016/j.ymben.2011.02.003
Kampranis SC, Ioannidis D, Purvis A, Mahrez W, Ninga E, Katerelos NA, Goodenough PW et al (2007) Rational conversion of substrate and product specificity in a Salvia monoterpene synthase: structural insights into the evolution of terpene synthase function. Plant Cell 19:1994–2005. https://doi.org/10.1105/tpc.106.047779
Karunanithi PS, Zerbe P (2019) Terpene synthases as metabolic gatekeepers in the evolution of plant terpenoid chemical diversity. Front Plant Sci 10:1166. https://doi.org/10.3389/fpls.2019.01166
Kasahara H, Hanada A, Kuzuyama T, Takagi M, Kamiya Y, Yamaguchi S (2002) Contribution of the mevalonate and methylerythritol phosphate pathways to the biosynthesis of gibberellins in Arabidopsis. J Biol Chem 277:45188–45194. https://doi.org/10.1074/jbc.M208659200
Katoh S, Hyatt D, Croteau R (2004) Altering product outcome in Abies grandis (−)limonene synthase and (−)-limonene/(−)-α-pinene synthase by domain swapping and directed mutagenesis. Arch Biochem Biophys 425:65–76. https://doi.org/10.1016/j.abb.2004.02.015
Keeling CI, Bohlmann J (2006) Genes, enzymes and chemicals of terpenoid diversity in the constitutive and induced defence of conifers against insects and pathogens. N Phytol 170:657–675. https://doi.org/10.1111/j.1469-8137.2006.01716.x
Keeling CI, Dullat HK, Yuen M, Ralph SG, Jancsik S (2010) Identification and functional characterization of monofunctional ent-copalyl diphosphate and ent-kaurene synthases in white spruce reveal different patterns for diterpene synthase evolution for primary and secondary metabolism in gymnosperms. Plant Physiol 152:1197–1208. https://doi.org/10.1104/pp.109.151456
Keeling CI, Weisshaar S, Ralph SG, Jancsik S, Hamberger B, Dullat HK (2011) Transcriptome mining, functional characterization, and phylogeny of a large terpene synthase gene family in spruce (Picea spp.). BMC Plant Biol 11:43. https://doi.org/10.1186/1471-2229-11-43
Kharel Y, Koyama T (2003) Molecular analysis of cis-prenyl chain elongating enzymes. Nat Prod Rep 20:111–118. https://doi.org/10.1039/B108934J
Kim YJ, Lee OR, Oh JY, Jang MG, Yang DC (2014) Functional analysis of 3-hydroxy-3-methylglutaryl coenzyme A reductase encoding genes in triterpene saponin-producing ginseng. Plant Physiol 165:373–387. https://doi.org/10.1104/pp.113.222596
Kirby J, Nishimoto M, Park JG, Withers ST, Nowroozi F, Behrendt D et al (2010) Cloning of casbene and neocembrene synthases from Euphorbiaceae plants and expression in Saccharomyces cerevisiae. Phytochemistry 71:1466–1473. https://doi.org/10.1016/j.phytochem.2010.06.001
Köksal M, Hu H, Coates RM, Peters RJ, Christianson DW (2011a) Structure and mechanism of the diterpene cyclase ent-copalyl diphosphate synthase. Nat Chem Biol 7:431. https://doi.org/10.1038/NChemBio.578
Köksal M, Jin Y, Coates RM, Croteau R, Christianson DW (2011b) Taxadiene synthase structure and evolution of modular architecture in terpene biosynthesis. Nature 469:116. https://doi.org/10.1038/nature09628
Köksal M, Potter K, Peters RJ, Christianson DW (2014) 1.55 Å-resolution structure of ent-copalyl diphosphate synthase and exploration of general acid function by site-directed mutagenesis. Biochim Biophys Acta 1840:184–190. https://doi.org/10.1016/j.bbagen.2013.09.004
Köpke D, Schröder R, Fischer HM, Gershenzon J, Hilker M, Schmidt A (2008) Does egg deposition by herbivorous pine sawflies affect transcription of sesquiterpene synthases in pine? Planta 228:427–438. https://doi.org/10.1007/s00425-008-0747-8
Külheim C, Padovan A, Hefer C, Krause ST, Köllner TG, Myburg AA et al (2015) The Eucalyptus terpene synthase gene family. BMC Genomics 16: https://doi.org/10.1186/s12864-015-1598-x
Kumar S, Hahn FM, Baidoo E, Kahlon TS, Wood DF, McMahan CM, Cornish K, Keasling JD, Daniell H, Whalen MC (2012) Remodeling the isoprenoid pathway in tobacco by expressing the cytoplasmic mevalonate pathway in chloroplasts. Metab Eng 14:19–28. https://doi.org/10.1016/j.ymben.2011.11.005
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. https://doi.org/10.1093/molbev/msy096
Laby RJ, Kincaid MS, Kim D, Gibson SI (2000) The Arabidopsis sugar-insensitive mutants sis4 and sis5 are defective in abscisic acid synthesis and response. Plant J 23:587–596. https://doi.org/10.1046/j.1365-313x.2000.00833.x
Lange I, Poirier BC, Herron BK, Lange BM (2015) Comprehensive assessment of transcriptional regulation facilitates metabolic engineering of isoprenoid accumulation in Arabidopsis. Plant Physiol 169:1595–1606. https://doi.org/10.1104/pp.15.00573
Laothawornkitkul J, Paul ND, Vickers CE, Possell M, Taylor JE, Mullineaux PM et al (2008) Isoprene emissions influence herbivore feeding decisions. Plant Cell Environ 31:1410–1415. https://doi.org/10.1111/j.1365-3040.2008.01849.x
Laule O, Furholz A, Chang HS, Zhu T, Wang X, Heifetz PB, Gruissem W, Lange M (2003) Crosstalk between cytosolic and plastidial pathways of isoprenoid biosynthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA 100:6866–6871. https://doi.org/10.1073/pnas.1031755100
Lerdau M, Gray D (2003) Ecology and evolution of light-dependent and light-independent phytogenic volatile organic carbon. N Phytol 157:199–211. https://doi.org/10.1046/j.1469-8137.2003.00673.x
Lesburg CA, Zhai G, Cane DE, Christianson DW (1997) Crystal structure of pentalenene synthase: mechanistic insights on terpenoid cyclization reactions in biology. Science 277:1820–1824. https://doi.org/10.1126/science.277.5333.1820
Li G, Köllner TG, Yin Y, Jiang Y, Xu Y, Gershenzon J et al (2012) Nonseed plant Selaginella moellendorffii has both seed plant and microbial types of terpene synthases. Proc Natl Acad Sci USA 109:14711–14715. https://doi.org/10.1073/pnas.1219604110
Loreto F, Fischbach RJ, Schnitzler JP, Ciccioli P, Brancaleoni E, Calfapietra C, Seufert G (2001) Monoterpene emission and monoterpene synthase activities in the Mediterranean evergreen oak Quercus ilex L. grown at elevated CO2 concentrations. Glob Change Biol 7:709–717. https://doi.org/10.1046/j.1354-1013.2001.00442.x
Loreto F, Dicke M, Schnitzler JP, Turlings TC (2014) Plant volatiles and the environment. Plant Cell Environ 37:1905–1908. https://doi.org/10.1111/pce.12369
Ma D, Li G, Zhu Y, Xie DY (2017) Overexpression and suppression of Artemisia annua 4-hydroxy-3-methylbut-2-enyl diphosphate reductase 1 gene (AaHDR1) differentially regulate artemisinin and terpenoid biosynthesis. Front Plant Sci 8:77. https://doi.org/10.3389/fpls.2017.00077
Mafu S, Hillwig ML, Peters RJ (2011) A novel labda-7, 13-E-dien-15-ol-producing bifunctional diterpene synthase from Selaginella moellendorffii. ChemBioChem 12:1984–1987. https://doi.org/10.1002/cbic.201100336
Martin DM, Fäldt J, Bohlmann J (2004) Functional characterization of nine Norway spruce TPS genes and evolution of gymnosperm terpene synthases of the TPS-d subfamily. Plant Physiol 135:1908–1927. https://doi.org/10.1104/pp.104.042028.1908
Martin DM, Aubourg S, Schouwey MB, Daviet L, Schalk M, Toub O et al (2010) Functional annotation, genome organization and phylogeny of the grapevine (Vitis vinifera) terpene synthase gene family based on genome assembly, FLcDNA cloning, and enzyme assays. BMC Plant Biol 10:226. https://doi.org/10.1186/1471-2229-10-226
Mau JD, West CA (1994) Cloning of casbene synthase cDNA: evidence for conserved structural features among terpenoid cyclases in plants. Proc Natl Acad Sci USA 91:8497–8501. https://doi.org/10.1073/pnas.91.18.8497
May B, Lange BM, Wüst M (2013) Biosynthesis of sesquiterpenes in grape berry exocarp of Vitis vinifera L.: evidence for a transport of farnesyl diphosphate precursors from plastids to the cytosol. Phytochemistry 95:135–144. https://doi.org/10.1016/j.phytochem.2013.07.021
Mcandrew RP, Peralta-Yahya PP, Degiovanni A, Pereira JH, Hadi MZ, Keasling JD, Adams PD (2011) Structure of a three-domain sesquiterpene synthase: a prospective target for advanced biofuels production. Structure 19:1876–1884. https://doi.org/10.1016/j.str.2011.09.013
Meier S, Tzfadia O, Vallabhaneni R, Gehring C, Wurtzel ET (2011) A transcriptional analysis of carotenoid, chlorophyll and plastidial isoprenoid biosynthesis genes during development and osmotic stress responses in Arabidopsis thaliana. BMC Syst Biol. https://doi.org/10.1186/1752-0509-5-77
Mendoza-Poudereux I, Kutzner E, Huber C, Segura J, Eisenreich W, Arrillaga I (2015) Metabolic cross-talk between pathways of terpenoid backbone biosynthesis in spike lavender. Plant Physiol Biochem 95:113–120. https://doi.org/10.1016/j.plaphy.2015.07.029
Mongelard G, Seemann M, Boisson AM, Rohmer M, Bligny R, Rivasseau C (2011) Measurement of carbon flux through the MEP pathway for isoprenoid synthesis by 31P-NMR spectroscopy after specific inhibition of 2-C-methyl-d-erythritol 2,4-cyclodiphosphate reductase.Effect of light and temperature. Plant Cell Environ 34:1241–1247. https://doi.org/10.1111/j.1365-3040.2011.02322.x
Monson RK, Jones RT, Rosenstiel TN, Schnitzler JP (2013) Why only some plants emit isoprene. Plant Cell Environ 36:503–516. https://doi.org/10.1111/pce.12015
Morrone D, Chambers J, Lowry L, Kim G, Anterola A, Bender K, Peters RJ (2009) Gibberellin biosynthesis in bacteria: separate ent-copalyl diphosphate and ent-kaurene synthases in Bradyrhizobium japonicum. FEBS Lett 583:475–480. https://doi.org/10.1016/j.febslet.2008.12.052
Mumm R, Posthumus MA, Dicke M (2008) Significance of terpenoids in induced indirect plant defence against herbivorous arthropods. Plant Cell Environ 31:575–585. https://doi.org/10.1111/j.1365-3040.2008.01783.x
Nagegowda DA, Gutensohn M, Wilkerson CG, Dudareva N, Lafayette W, Consortium MP, Lansing E (2008) Two nearly identical terpene synthases catalyze the formation of nerolidol and linalool in snapdragon flowers. Plant J 55:224–239. https://doi.org/10.1111/j.1365-313X.2008.03496.x
Neale DB, Wegrzyn JL, Stevens KA, Zimin AV, Puiu D, Crepeau MW et al (2014) Decoding the massive genome of loblolly pine using haploid DNA and novel assembly strategies. Genome Biol 15:R59. https://doi.org/10.1186/gb-2014-15-3-r59
Nelson DR (2011) Progress in tracing the evolutionary paths of cytochrome P450. Biochim Biophys Acta Proteins Proteomics 1814:14–18. https://doi.org/10.1016/j.bbapap.2010.08.008
Nicolaci A, Travaglini D, Menguzzato G, Nocentini S, Veltri A, Iovino F (2014) Ecological and anthropogenic drivers of Calabrian pine (Pinus nigra JF Arn. ssp. laricio (Poiret) Maire) distribution in the Sila Mountain Range. iForest 8:497. https://doi.org/10.3832/ifor1041-007
Nystedt B, Street NR, Wetterbom A, Zuccolo A, Lin YC, Scofield DG et al (2013) The Norway spruce genome sequence and conifer genome evolution. Nature 497:579–584. https://doi.org/10.1038/nature12211
Opitz S, Nes WD, Gershenzon J (2014) Both methylerythritol phosphate and mevalonate pathways contribute to biosynthesis of each of the major isoprenoid classes in young cotton seedlings. Phytochemistry 98:110–119. https://doi.org/10.1016/j.phytochem.2013.11.010
Orlova I, Varbanova M, Fridman E, Yamaguchi S, Hanada A, Kamiya Y, Krichevsky A (2009) The small subunit of snapdragon geranyl diphosphate synthase modifies the chain length specificity of tobacco geranylgeranyl diphosphate synthase in planta. Plant Cell 21:4002–4017. https://doi.org/10.1105/tpc.109.071282
Pazouki L, Niinemets Ü (2016) Multi-substrate terpene synthases: their occurrence and physiological significance. Front Plant Sci 7:1019. https://doi.org/10.3389/fpls.2016.01019
Peters RJ (2006) Uncovering the complex metabolic network underlying diterpenoid phytoalexin biosynthesis in rice and other cereal crop plants. Phytochemistry 67:2307–2317. https://doi.org/10.1016/j.phytochem.2006.08.009
Peters RJ, Flory JE, Jetter R, Ravn MM, Lee HJ, Coates RM, Croteau RB (2000) Abietadiene synthase from grand fir (Abies grandis): characterization and mechanism of action of the “pseudomature” recombinant enzyme. Biochemistry 39:15592–15602. https://doi.org/10.1021/bi001997l
Peters RJ, Carter OA, Zhang Y, Matthews BW, Croteau RB (2003) Bifunctional abietadiene synthase: mutual structural dependence of the active sites for protonation-initiated and ionization-initiated cyclizations. Biochemistry 42:2700–2707. https://doi.org/10.1021/bi020492n
Phillips MA, Wildung MR, Williams DC, Hyatt DC, Croteau R (2003) cDNA isolation, functional expression, and characterization of (+)-α-pinene synthase and (−)-α-pinene synthase from loblolly pine (Pinus taeda): stereocontrol in pinene biosynthesis. Arch Biochem Biophys 411:267–276. https://doi.org/10.1016/S0003-9861(02)00746-4
Pierik R, Ballaré CL, Dicke M (2014) Ecology of plant volatiles: taking a plant community perspective. Plant Cell Environ 37:1845–1853. https://doi.org/10.1111/pce.12330
Polge C, Thomas M (2007) SNF1/AMPK/SnRK1 kinases, global regulators at the heart of energy control? Trends Plant Sci 12:20–28. https://doi.org/10.1016/j.tplants.2006.11.005
Prisic S, Xu M, Wilderman PR, Peters RJ (2004) Rice contains two disparate ent-copalyl diphosphate synthases with distinct metabolic functions. Plant Physiol 136:4228–4236. https://doi.org/10.1104/pp.104.050567.4228
Rajabi MH, Pazouki L, Niinemets Ü (2013) The biochemistry and molecular biology of volatile messengers in trees. Biology, controls and models of tree volatile organic compound emissions. Springer, Dordrecht, pp 47–93
Rasmann S, Köllner TG, Degenhardt J, Hiltpold I, Toepfer S, Kuhlmann U et al (2005) Recruitment of entomopathogenic nematodes by insect-damaged maize roots. Nature 434:732. https://doi.org/10.1038/nature03451DO
Rasulov B, Talts E, Kännaste A, Niinemets Ü (2015) Bisphosphonate inhibitors reveal a large elasticity of plastidic isoprenoid synthesis pathway in isoprene-emitting hybrid aspen. Plant Physiol 168:532–548. https://doi.org/10.1104/pp.15.00470
Ro DK, Bohlmann J (2006) Diterpene resin acid biosynthesis in loblolly pine (Pinus taeda): functional characterization of abietadiene/levopimaradiene synthase (PtTPS-LAS) cDNA and subcellular targeting of PtTPS-LAS and abietadienol/abietadienal oxidase (PtAO, CYP720B1). Phytochemistry 67:1572–1578. https://doi.org/10.1016/j.phytochem.2006.01.011
Ro DK, Arimura GI, Lau SY, Piers E, Bohlmann J (2005) Loblolly pine abietadienol/abietadienal oxidase PtAO (CYP720B1) is a multifunctional, multisubstrate cytochrome P450 monooxygenase. Proc Natl Acad Sci USA 102:8060–8065. https://doi.org/10.1073/pnas.0500825102
Rodríguez-Concepción M (2006) Early steps in isoprenoid biosynthesis: multilevel regulation of the supply of common precursors in plant cells. Phytochem Rev 5:1–15. https://doi.org/10.1007/s11101-005-3130-4
Rodríguez-Concepción M, Boronat A (2015) Breaking new ground in the regulation of the early steps of plant isoprenoid biosynthesis. Curr Opin Plant Biol 25:17–22. https://doi.org/10.1016/j.pbi.2015.04.001
Rodríguez-Concepción M, Fores O, Martinez-Garcia JF, Gonzalez V, Phillips MA, Ferrer A, Boronat A (2004) Distinct light-mediated pathways regulate the biosynthesis and exchange of isoprenoid precursors during Arabidopsis seedling development. Plant Cell 16:144–156. https://doi.org/10.1105/tpc.016204
Sallaud C, Rontein D, Onillon S, Jabès F, Duffé P, Giacalone C et al (2009) A novel pathway for sesquiterpene biosynthesis from Z, Z-farnesyl pyrophosphate in the wild tomato Solanum habrochaites. Plant Cell 21:301–317. https://doi.org/10.1105/tpc.107.057885
Schilmiller AL, Schauvinhold I, Larson M, Xu R, Charbonneau AL, Schmidt A et al (2009) Monoterpenes in the glandular trichomes of tomato are synthesized from a neryl diphosphate precursor rather than geranyl diphosphate. Proc Natl Acad Sci USA 106:10865–10870. https://doi.org/10.1073/pnas.0904113106
Schnitzler JP, Louis S, Behnke K, Loivamäki M (2010) Poplar volatiles—biosynthesis, regulation and (eco) physiology of isoprene and stress-induced isoprenoids. Plant Biol 12:302–316. https://doi.org/10.1111/j.1438-8677.2009.00284.x
Shalev TJ, Yuen MM, Gesell A, Yuen A, Russell JH, Bohlmann J (2018) An annotated transcriptome of highly inbred Thuja plicata (Cupressaceae) and its utility for gene discovery of terpenoid biosynthesis and conifer defense. Tree Genet Genomes 14:35. https://doi.org/10.1007/s11295-018-1248-y
Sharkey TD, Yeh S (2001) Isoprene emission from plants. Annu Rev Plant Biol 52:407–436. https://doi.org/10.1146/annurev.arplant.52.1.407
Sharkey TD, Gray DW, Pell HK, Breneman SR, Topper L (2013) Isoprene synthase genes form a monophyletic clade of acyclic terpene synthases in the tps-b terpene synthase family. Evolution 67:1026–1040. https://doi.org/10.1111/evo.12013
Shi M, Luo X, Ju G, Yu X, Hao X, Huang Q, Xiao J, Cui L, Kai G (2014) Increased accumulation of the cardio-cerebrovascular disease treatment drug Tanshinone in Salvia miltiorrhiza hairy roots by the enzymes 3-hydroxy-3-methylglutaryl CoA reductase and 1-deoxy-d-xylulose 5-phosphate reductoisomerase. Funct Integr Genomics 14:603–615. https://doi.org/10.1007/s10142-014-0385-0
Singh B, Sharma RA (2015). Plant terpenes: defense responses, phylogenetic analysis, regulation and clinical applications. 3 Biotech 5:129–151. https://doi.org/10.1007/s13205-014-0220-2
Surmacz L, Swiezewska E (2011) Biochemical and biophysical research communications polyisoprenoids—secondary metabolites or physiologically important superlipids? Biochem Biophys Res Commun 407:627–632. https://doi.org/10.1016/j.bbrc.2011.03.059
Takahashi S, Koyama T (2006) Structure and function of cis-prenyl chain elongating enzymes. Chem Rec 6:194–205. https://doi.org/10.1002/tcr.20083
Tarshis LC, Proteau PJ, Kellogg BA, Sacchettini JC, Poulter CD (1996) Regulation of product chain length by isoprenyl diphosphate synthases. Proc Natl Acad Sci USA 93:15018–15023. https://doi.org/10.1073/pnas.93.26.15018
Thimmappa R, Geisler K, Louveau T, Maille PO, Osbourn A (2014) Triterpene biosynthesis in plants. Annu Rev Plant Biol 65:225–257. https://doi.org/10.1146/annurev-arplant-050312-120229
Tholl D (2015) Biosynthesis and biological functions of terpenoids in plants. Biotechnology of isoprenoids. Springer, Cham, pp 63–106
Tholl D, Lee S (2011) Terpene specialized metabolism in Arabidopsis thaliana. Arabidopsis Book 9:e0143. https://doi.org/10.1043/tab.0143
Townsend BJ, Poole A, Blake CJ, Llewellyn DJ (2005) Antisense suppression of a (+)-δ-cadinene synthase gene in cotton prevents the induction of this defense response gene during bacterial blight infection but not its constitutive expression. Plant Physiol 138:516–528. https://doi.org/10.1104/pp.104.056010
Trapp SC, Croteau RB (2001) Genomic organization of plant terpene synthases and molecular evolutionary implications. Genetics 158:811–832
Vannice JC, Skaff DA, Keightley A, Addo JK, Wyckoff GJ, Miziorko HM (2014) Identification in Haloferax volcanii of phosphomevalonate decarboxylase and isopentenyl phosphate kinase as catalysts of the terminal enzyme reactions in an archaeal alternate mevalonate pathway. J Bacteriol 196:1055–1063. https://doi.org/10.1128/JB.01230-13
Vaughan MM, Wang Q, Webster FX, Kiemle D, Hong YJ, Tantillo DJ et al (2013) Formation of the unusual semivolatile diterpene rhizathalene by the Arabidopsis class I terpene synthase TPS08 in the root stele is involved in defense against belowground herbivory. Plant Cell 25:1108–1125. https://doi.org/10.1105/tpc.112.100057
Velikova V, Ghirardo A, Vanzo E, Merl J, Hauck SM, Schnitzler JP (2014) Genetic manipulation of isoprene emissions in poplar plants remodels the chloroplast proteome. J Proteome Res 13:2005–2018
Vranova E, Coman D, Gruissem W (2013) Network analysis of the MVA and MEP pathways for isoprenoid synthesis. Annu Rev Plant Biol 64:665–700. https://doi.org/10.1146/annurev-arplant-050312-120116
Warren RL, Keeling CI, Yuen MM, Raymond A, Taylor GA, Vandervalk BP et al (2015) Improved white spruce (Picea glauca) genome assemblies and annotation of large gene families of conifer terpenoid and phenolic defense metabolism. Plant J 83:189–212. https://doi.org/10.1111/tpj.12886
Whittington DA, Wise ML, Urbansky M, Coates RM, Croteau RB, Christianson DW (2002) Bornyl diphosphate synthase: structure and strategy for carbocation manipulation by a terpenoid cyclase. Proc Natl Acad Sci USA 99:15375–15380. https://doi.org/10.1073/pnas.232591099
Wildung MR, Croteau R (1996) A cDNA clone for taxadiene synthase, the diterpene cyclase that catalyzes the committed step of taxol biosynthesis. J Biol Chem 27:9201–9204. https://doi.org/10.1074/jbc.271.16.9201
Wright GA, Schiestl FP (2009) The evolution of floral scent: the influence of olfactory learning by insect pollinators on the honest signalling of floral rewards. Funct Ecol 23:841–851. https://doi.org/10.1111/j.1365-2435.2009.01627.x
Xu M, Wilderman PR, Morrone D, Xu J, Roy A, Margis-Pinheiro M et al (2007) Functional characterization of the rice kaurene synthase-like gene family. Phytochemistry 68:312–326. https://doi.org/10.1016/j.phytochem.2006.10.016
Xu M, Galhano R, Wiemann P, Bueno E, Tiernan M, Wu W, Peters RJ et al (2012) Genetic evidence for natural product-mediated plant–plant allelopathy in rice (Oryza sativa). N Phytol 193:570–575. https://doi.org/10.1111/j.1469-8137.2011.04005.x
Zerbe P, Chiang A, Yuen M, Hamberger B, Draper JA, Bohlmann J et al (2012) Bifunctional cis-abienol synthase from Abies balsamea discovered by transcriptome sequencing and its implications for diterpenoid fragrance production. J Biol Chem 287:12121–12131. https://doi.org/10.1074/jbc.M111.317669
Zerbe P, Hamberger B, Yuen MM, Chiang A, Sandhu HK, Madilao LL et al (2013) Gene discovery of modular diterpene metabolism in nonmodel systems. Plant Physiol 162:1073–1091. https://doi.org/10.1104/pp.113.218347
Zhou K, Gao Y, Hoy JA, Mann FM, Honzatko RB, Peters RJ (2012) Insights into diterpene cyclization from structure of bifunctional abietadiene synthase from Abies grandis. J Biol Chem 287:6840–6850. https://doi.org/10.1074/jbc.M111.337592
Zi J, Matsuba Y, Hong YJ, Jackson AJ, Tantillo DJ, Pichersky E, Peters RJ (2014) Biosynthesis of lycosantalonol, a cis-prenyl derived diterpenoid. J Am Chem Soc 136:16951–16953. https://doi.org/10.1021/ja508477e
Zulak KG, Bohlmann J (2010) Terpenoid biosynthesis and specialized vascular cells of conifer defense. J Integr Plant Biol 52:86–97. https://doi.org/10.1111/j.1744-7909.2010.00910.x
Funding
The present work was carried out in the framework of the “ALForLab” Project (PON03PE_00024_1), co-funded by the National Operational Programme for Research and Competitiveness (PON R&C) 2007–2013, through the European Regional Development Fund (ERDF) and National Resource (Revolving Fund-Cohesion Action Plan (CAP) MIUR).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Handling editor: Carrie Olson-Manning.
Electronic supplementary material
Below is the link to the electronic supplementary material.
239_2020_9930_MOESM1_ESM.pdf
Supplementary file1 (PDF 510 kb) Table S1 Full-length cDNA sequences retrieved from the NCBI database upon which the phylogenetic analysis of terpene synthases in gymnosperms was carried out (Fig. 5). The ent-kaurene synthase from the moss Physcomitrella patens was included as outgroup
239_2020_9930_MOESM2_ESM.pdf
Supplementary file2 (PDF 277 kb) Table S2 Full-length cDNA sequences of functionally characterized terpene synthases (TPSs) employed for the BLAST search in the NCBI database of the putative TPSs of Pinus spp.
239_2020_9930_MOESM3_ESM.tiff
Supplementary file3 (TIFF 1751 kb) Fig. S1 Phylogenetic tree for the deduced amino acid sequences of the 2-methyl-3-buten-2-ol synthases (MBOSs) from Pinus species identified in NCBI database (Table 3). Physcomitrella patens ent-kaurene synthase (Pt TPS-entKS) was used to root the tree
239_2020_9930_MOESM4_ESM.tiff
Supplementary file4 (TIFF 3159 kb) Fig. S2 Strategy adopted for the genomic amplification of a putative Pinus nigra subsp. laricio gene belonging to the phylogenetic TPS-d1 Group 3. a Schematic representation of the full-length cDNA of a representative member of the phylogenetic TPS-d1 Group 3 (PcMTPS4 from Pinus contorta, in the present case, see Fig. 6; Table 3) in which the positions of the forward (F2) and the reverse (R3) primers used in the amplification of genomic DNA are shown. b Intron (yellow)/exon (blue) structure of the amplified Pinus nigra subsp. laricio genomic sequence. The positions of the primers used to amplify the genomic fragment are also shown
239_2020_9930_MOESM5_ESM.tiff
Supplementary file5 (TIFF 4184 kb) Fig. S3 Phylogenetic tree of the deduced amino acid sequences obtained by combining monoterpene synthases (MTPSs) and the five selected 2-methyl-3-buten-2-ol synthases (MBOSs) identified in different Pinus species (Fig. 6; Table 3) and the six sequences isolated from Pinus nigra subsp. laricio (outlined in red). Physcomitrella patens ent-kaurene synthase (Pt TPS-entKS) was used to root the tree. Branches indicated with dots represent bootstrap support more than 80% (100 repetitions). The seven phylogenetic groups identified in the pine members of TPS-d1 clade are highlighted with square brackets. For acronyms denoting plants species, see Table 3
239_2020_9930_MOESM6_ESM.pdf
Supplementary file6 (PDF 324 kb) Experimental procedures: Notes: Data archiving statement The cDNA and genomic sequences of the monoterpene synthases (MTPSs) from Pinus nigra subsp. laricio will appear in the GenBank database with the following accession numbers: MN088807 (Pnl_MBOS_1.1), MN088808 (Pnl_MTPS_1.2), MN088809 (Pnl_MTPS_1.5), MN088810 (Pnl_MTPS_1.7), MN088811 (Pnl_MTPS_1.4) and MN088812 (Pnl_MTPS_1.3)
Rights and permissions
About this article
Cite this article
Alicandri, E., Paolacci, A.R., Osadolor, S. et al. On the Evolution and Functional Diversity of Terpene Synthases in the Pinus Species: A Review. J Mol Evol 88, 253–283 (2020). https://doi.org/10.1007/s00239-020-09930-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-020-09930-8