Abstract
Cycles of biologically relevant reactions are an alternative to an origin of life emerging from a steady state away from equilibrium. The cycles involve a rate at which polymers are synthesized and accumulate in microscopic compartments called protocells, and two rates in which monomers and polymers are chemically degraded by hydrolytic reactions. Recent experiments have demonstrated that polymers are synthesized from mononucleotides and accumulate during cycles of hydration and dehydration, which means that the rate of polymer synthesis during the dehydrated phase of the cycle is balanced (but not dominated) by the rate of polymer hydrolysis during the hydrated phase of the cycle. Furthermore, depurination must be balanced by the reverse process of repurination. Here we describe a computational model that was inspired by experimental results, can be generalized to accommodate other reaction parameters, and has qualitative predictive power.






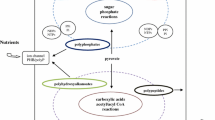
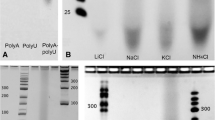
(Reproduced with permission from Da Silva et al. (2015))

(a Reproduced with permission from DeGuzman et al. (2014))
Similar content being viewed by others
Notes
The choice of a 2-min wet phase and 30-min dry phase in a cycle is based on previous laboratory studies. These durations are reasonable because the water levels of hydrothermal pools undergo periodic fluctuations related to precipitation and evaporation as well as geyser activity. A longer dry phase is required for condensation reactions leading to polymerization, while the wet phase must not be so long that the hydrolysis of polymers dominates over the synthesis of polymers.
Abbreviations
- \(s_n\) :
-
A vector describing the state of the system after an n number of cycles
- \(P_n\) :
-
The mass of intact RNA polymer in the system after an n number of cycles
- \(\pi _n\) :
-
The mass of depurinated RNA polymer in the system after an n number of cycles
- \(M_n\) :
-
The mass of intact RNA monomer in the system after an n number of cycles
- \(\mu _n\) :
-
The mass of depurinated RNA monomer in the system after an n-number of cycles
- C :
-
The matrix that describes the transformation applied to the system over the course of one cycle
- h :
-
The proportion of polymer mass converted to monomer mass per cycle by the cleavage of phosphodiester bonds during the wet phase
- m :
-
The proportion of monomer mass converted to polymer mass per cycle by the formation of phosphodiester bonds during the dry phase
- \(d_{\text{m}}\) :
-
The proportion of monomer mass depurinated per cycle during the wet phase
- \(d_{\text{p}}\) :
-
The proportion of polymer mass depurinated per cycle during the wet phase
- \(r_{\text{m}}\) :
-
The proportion of degraded monomer mass repurinated per cycle during the dry phase
- \(r_{\text{p}}\) :
-
The proportion of degraded polymer mass repurinated per cycle during the dry phase
- D :
-
The matrix that describes the transformation applied to the system over the course of the dry phase of one cycle
- W :
-
The matrix that describes the transformation applied to the system over the course of the wet phase of one cycle
- n :
-
The n number of cycles after which the mass of depurinated (non-reactive) RNA monomer has accumulated
References
Da Silva L, Maurel MC, Deamer D (2015) Salt-promoted synthesis of RNA-like molecules in simulated hydrothermal conditions. J Mol Evol 80:86–97
Damer B, Deamer D (2015) Coupled phases and combinatorial selection in fluctuating hydrothermal pools: a scenario to guide experimental approaches to the origin of cellular life. Life 5:872–887
DeGuzman V, Vercoutere W, Shenasa H, Deamer D (2014) Generation of oligonucleotides under hydrothermal conditions by non-enzymatic polymerization. J Mol Evol 78:251–262
Higgs PG (2016) The effect of limited diffusion and wet-dry cycling on reversible polymerization reactions: implications for prebiotic synthesis of nucleic acids. Life. https://doi.org/10.3390/life6020024
Lindahl T (1993) Instability and decay of the primary structure of DNA. Nature 362:709–715
Lorig-Roach R, Deamer D (2018) Condensation and decomposition of nucleotides in simulated hydrothermal fields. In: Menor-Salvan C (ed) Prebiotic chemistry and chemical evolution of the nucleic acids. Springer, New York
Ma W, Yu C, Zhang W (2007a) Monte Carlo simulation of early molecular evolution in the RNA world. Biosystems 90:28–39
Ma W, Yu C, Zhang W, Hu J (2007b) Nucleotide synthetase ribozymes may have emerged first in the RNA world. RNA 13:2012–2019
Ma W, Yu C, Zhang W, Zhou P, Hu J (2011) Self-replication: spelling it out in a chemical background. Theory Biosci 130:119–125
Nam I, Nam HG, Zare RN (2018) Abiotic synthesis of purine and pyrimidine ribonucleosides in aqueous microdroplets. Proc Natl Acad Sci USA 115:36–40
Pearce BKD, Pudritz RE, Semenov DA, Henning TK (2017) Origin of the RNA world: the fate of nucleobases in warm little ponds. Proc Natl Acad Sci USA 114:11327–11332
Rajamani S, Vlassov A, Benner S, Coombs A, Olasagasti F, Deamer D (2008) Lipid-assisted synthesis of RNA-like polymers from mononucleotides. Orig Life Evol Biosph 38:57–74
Ross DS, Deamer D (2016) Dry/wet cycling and the thermodynamics and kinetics of prebiotic polymer synthesis. Life. https://doi.org/10.3390/life6030028
Walker SI, Grover MA, Hud NV (2012) Universal sequence replication, reversible polymerization and early functional biopolymers: a model for the initiation of prebiotic sequence evolution. PLoS ONE 7:1–15
Acknowledgements
We would like to thank Paul Higgs and David Ross for valuable discussion of the manuscript. Further, we would like to thank Gilbert Strang for inspiring us to take a linear algebraic approach to the analysis of these processes. Finally, we would like to thank the referees for taking the time to review and provide their council regarding this research.
Funding
The funding was provided by Hierarchical Systems Foundation.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Hargrave, M., Thompson, S.K. & Deamer, D. Computational Models of Polymer Synthesis Driven by Dehydration/Rehydration Cycles: Repurination in Simulated Hydrothermal Fields. J Mol Evol 86, 501–510 (2018). https://doi.org/10.1007/s00239-018-9865-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-018-9865-5