Abstract
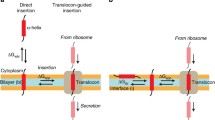
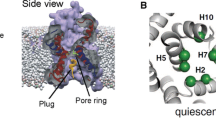
The cellular translocon, present in all three domains of life, is one of the most versatile and important biological nanopores. This complex molecular apparatus is directly responsible for the secretion of globular proteins across membranes as well as the insertion of integral membrane proteins into lipid bilayers. Recently determined structures of the archaean SecY translocon reveal an hour-glass-shaped pore, which accommodates the nascent peptide chain during translocation. While these structures provide important insights into ribosome binding to the translocon, threading of the nascent chain into the channel, and lateral gate opening for releasing the folded helical peptide into the membrane bilayer, the exact folding pathway of the peptide inside the protein-conducting channel during translocation and prior to the lateral release into the bilayer remains elusive. In the present study, we use molecular dynamics simulations to investigate atomic resolution peptide folding in hour-glass-shaped pore models that are based on the SecY translocon channel structure. The theoretical setup allows systematic variation of key determinants of folding, in particular the degree of confinement of the peptide and the hydration level of the pore. A 27-residue hydrophobic peptide was studied that is preferentially inserted into membranes by the translocon. Our results show that both pore diameter as well as channel hydration are important determinants for folding efficiency and helical stability of the peptide, therefore providing important insights into translocon gating and lateral peptide partitioning.







Similar content being viewed by others
References
Andersen HC (1983) Rattle: a “velocity” version of the shake algorithm for molecular dynamics calculations. J Comput Phys 52:24–34
Anishkin A, Sukharev S (2004) Water dynamics and dewetting transitions in the small mechanosensitive channel MscS. Biophys J 86:2883–2895
Beckstein O, Sansom MSP (2003) Liquid-vapor oscillations of water in hydrophobic nanopores. Proc Natl Acad Sci USA 100:7063–7068
Beckstein O, Sansom MSP (2004) The influence of geometry, surface character, and flexibility on the permeation of ions and water through biological pores. Phys Biol 1:42–52
Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, Haak JR (1984) Molecular dynamics with coupling to an external bath. J Chem Phys 81:3684–3690
Cheng Z, Gilmore R (2006) Slow translocon gating causes cytosolic exposure of transmembrane and lumenal domains during membrane protein integration. Nat Struct Mol Biol 13:930–936
Curnow P, Booth PJ (2009) The transition state for integral membrane protein folding. Proc Natl Acad Sci USA 106:773–778
Cymer F, von Heijne G (2013) Cotranslational folding of membrane proteins probed by arrest-peptide-mediated force measurements. Proc Natl Acad Sci USA 110:14640–14645
Cymer F, von Heijne G, White SH (2014) Mechanisms of integral membrane protein insertion and folding. J Mol Biol 427:999–1022
Frauenfeld J, Gumbart J, Sluis EO, Funes S, Gartmann M, Beatrix B, Mielke T, Berninghausen O, Becker T, Schulten K, Beckmann R (2011) Cryo-EM structure of the ribosome-SecYE complex in the membrane environment. Nat Struct Mol Biol 18:614–621
Gumbart J, Schulten K (2006) Molecular dynamics studies of the archaeal translocon. Biophys J 90:2356–2367
Gumbart J, Schulten K (2007) Structural determinants of lateral gate opening in the protein translocon. Biochemistry 46:11147–11157
Gumbart J, Schulten K (2008) The roles of pore ring and plug in the SecY protein-conducting channel. J Gen Physiol 132:709–719
Gumbart J, Chipot C, Schulten K (2011) Free-energy cost for translocon-assisted insertion of membrane proteins. Proc Natl Acad Sci USA 108:3596–3601
Gumbart JC, Teo I, Roux B, Schulten K (2013) Reconciling the roles of kinetic and thermodynamic factors in membrane-protein insertion. J Am Chem Soc 135:2291–2297
Haider S, Hall BA, Sansom MSP (2006) Simulations of a protein translocation pore: SecY. Biochemistry 45:13018–13024
Hershey JW (1991) Translational control in mammalian cells. Annu Rev Biochem 60:717–755
Hess B, Bekker H, Berendsen HJC, Fraaije JGEM (1997) LINCS: a linear constraint solver for molecular simulations. J Comput Chem 18:1463–1472
Hess B, Kutzner C, van der Spoel D, Lindahl E (2008) GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput 4:435–447
Hessa T, Kim H, Bihlmaier K, Lundin C, Boekel J, Andersson H, Nilsson I, White SH, von Heijne G (2005) Recognition of transmembrane helices by the endoplasmic reticulum translocon. Nature 433:377–381
Hessa T, Meindl-Beinker NM, Bernsel A, Kim H, Sato Y, Lerch-Bader M, Nilsson I, White SH, von Heijne G (2007) Molecular code for transmembrane-helix recognition by the Sec61 translocon. Nature 450:1026–1030
Humphrey W, Dalke A, Schulten K (1996) VMD—visual molecular dynamics. J Mol Graph 14:33–38
Ismail N, Hedman R, Linden M, von Heijne G (2015) Charge-driven dynamics of nascent-chain movement through the SecYEG translocon. Nat Struct Mol Biol 22(2):145–149
Jacobs RE, White SH (1989) The nature of the hydrophobic binding of small peptides at the bilayer interface: implications for the insertion of transbilayer helices. Biochemistry 28:3421–3437
Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML (1983) Comparison of simple potential functions for simulating liquid water. J Chem Phys 79:926–935
Jorgensen WL, Maxwell DS, Tirado-Rives J (1996) Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J Am Chem Soc 118:11225–11236
Lucent D, Vishal V, Pande VS (2007) Protein folding under confinement: a role for solvent. Proc Natl Acad Sci USA 104:10430–10434
Ojeda PA, Londono A, Nan-Yow C, Garcia M (2009) Monte Carlo simulations of proteins in cages: influence of confinement on the stability of intermediate states. Biophys J 96:1076–1082
Park E, Menetret JF, Gumbart JC, Ludtke SJ, Li W, Whynot A, Rapoport TA, Akey CW (2014) Structure of the SecY channel during initiation of protein translocation. Nature 506:102–106
Popot JL, Engelman DM (1990) Membrane-protein folding and oligomerization—the 2-stage model. Biochemistry 29:4031–4037
Rapoport TA (2007) Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature 450:663–669
Smart OS, Neduvelil JG, Wang X, Wallace BA, Sansom MSP (1996) HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J Mol Graph Model 14:354
Sorin EJ, Pande VS (2006) Nanotube confinement denatures protein helices. J Am Chem Soc 128:6316–6317
Takagi F, Koga N, Takada S (2003) How protein thermodynamics and folding mechanisms are altered by the chaperonin cage: molecular simulations. Proc Natl Acad Sci USA 100:11367–11372
Ulmschneider JP, Ulmschneider MB (2008a) Sampling efficiency in explicit and implicit membrane environments studied by peptide folding simulations. Proteins 75(3):586–597
Ulmschneider MB, Ulmschneider JP (2008b) Membrane adsorption, folding, insertion and translocation of synthetic trans-membrane peptides. Mol Membr Biol 25:245–257
Ulmschneider JP, Ulmschneider MB, Di Nola A (2006a) Monte Carlo vs molecular dynamics for all-atom polypeptide folding simulations. J Phys Chem B 110:16733–16742
Ulmschneider MB, Sansom MS, Di Nola A (2006b) Evaluating tilt angles of membrane-associated helices: comparison of computational and NMR techniques. Biophys J 90:1650–1660
Ulmschneider MB, Ulmschneider JP, Sansom MSP, Di Nola A (2007) A generalized born implicit membrane representation compared to experimental insertion free energies. Biophys J 92:2338–2349
Ulmschneider MB, Doux JPF, Killian JA, Smith J, Ulmschneider JP (2010) Mechanism and kinetics of peptide partitioning into membranes. J Am Chem Soc 132:3452–3460
Van den Berg B, Clemons WM Jr, Collinson I, Modis Y, Hartmann E, Harrison SC, Rapoport TA (2004) X-ray structure of a protein-conducting channel. Nature 427:36–44
Voorhees RM, Fernandez IS, Scheres SH, Hegde RS (2014) Structure of the mammalian ribosome-Sec61 complex to 3.4 A resolution. Cell 157:1632–1643
White SH, von Heijne G (2008) How translocons select transmembrane helices. Ann Rev Biophys 37:23–42
White SH, Wimley WC (1999) Membrane protein folding and stability: physical principles. Annu Rev Biophys Biomol Struct 28:319–365
Zhang B, Miller TF 3rd (2012a) Direct simulation of early-stage Sec-facilitated protein translocation. J Am Chem Soc 134:13700–13707
Zhang B, Miller TF 3rd (2012b) Long-timescale dynamics and regulation of Sec-facilitated protein translocation. Cell Rep 2:927–937
Zhou HX, Dill KA (2001) Stabilization of proteins in confined spaces. Biochemistry 40:11289–11293
Zimmer J, Nam Y, Rapoport TA (2008) Structure of a complex of the ATPase SecA and the protein-translocation channel. Nature 455:936–943
Ziv G, Haran G, Thirumalai D (2005) Ribosome exit tunnel can entropically stabilize α-helices. Proc Natl Acad Sci USA 102:18956–18961
Acknowledgments
Images were prepared with VMD and rendered with Tachyon (Humphrey et al. 1996).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ulmschneider, M.B., Koehler Leman, J., Fennell, H. et al. Peptide Folding in Translocon-Like Pores. J Membrane Biol 248, 407–417 (2015). https://doi.org/10.1007/s00232-015-9808-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00232-015-9808-7