Abstract
Sacoglossa are known for stealing photosynthetically active chloroplasts from their macroalgal food and incorporating them into their cytosol. The nutritional support these alien organelles (kleptoplasts) provide to the slugs is still debatable. Comparing slugs starved in continuous darkness (non-photosynthetic condition) and light (photosynthetic condition) is often used to understand the contribution of the kleptoplasts to the slugs' metabolism. Here, we examined the slugs' side of starvation in darkness to better understand the effects of darkness on the slugs. We compared the gene expression profile and digestive activity of Elysia viridis, starved for one week under ambient light and continuous darkness. Starvation in darkness led to the up-regulation of genes related to glucose deficiency, while genes involved in the development, cellular organization, and reproduction were down-regulated. This specific gene expression may counteract reduced nutrient availability under non-photosynthetic conditions. Under photosynthetic conditions, kleptoplasts may have a higher nutritional value and may be able to support some metabolic processes. It appears that the slugs can only access kleptoplast photosynthates through autophagy during starvation. Nevertheless, autophagy and length reduction in darkness are highly elevated compared to light conditions, suggesting that more slug tissue is needed to satisfy the nutritional demands under non-photosynthetic conditions. Since we did not detect a gene expression related to the export of photosynthates to the slugs, our results support the hypothesis that slugs use kleptoplasts as larders accessible via autophagy. As long as the kleptoplasts are functional, they provide an energetic support, helping the slugs to reduce starvation-induced stress.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The symbiotic relationship between animals and photosynthetic organisms is known as photosymbiosis and is considered especially beneficial for animal hosts (Melo Clavijo et al. 2018). Corals and their symbionts, the unicellular algae Symbiodiniaceae, are one of the best-studied systems of photosymbioses. The nutritional boost provided by symbionts increases the calcification rate of corals, facilitating the formation of large reef structures (Stanley and Lipps 2011). However, there is a special form of photosymbiosis found in many sacoglossan sea slugs (Händeler et al. 2009) and a few microturbellarian flatworms (Van Steenkiste et al. 2019). These two taxa only incorporate the chloroplasts from their algal prey into their cells, a process known as kleptoplasty. In some Sacoglossa, the “stolen” chloroplasts (kleptoplasts) remain photosynthetically active for weeks or months in the absence of any support from the algal nucleus (Wägele and Martin 2014). Even after decades of research, it is still uncertain what mechanisms are necessary to enable this so-called functional kleptoplasty in Sacoglossa. In the slugs, the expression of genes involved in kleptoplast recognition (Melo Clavijo et al. 2020; Mendoza et al. 2022) and maintenance (de Vries et al. 2015; Chan et al. 2018; Maeda et al. 2021) may mediate the establishment of functional kleptoplasty. Furthermore, the phototactic behavior of the slugs (Weaver and Clark 1981; Schmitt and Wägele 2011; Cartaxana et al. 2018) and chloroplast photoprotection mechanisms (Christa et al. 2018; Cartaxana et al. 2019; Havurinne and Tyystjärvi 2020; Havurinne et al. 2021) are considered to increase the longevity of kleptoplasts in slugs.
Compared to other photosymbiotic systems, the nutritional relevance of bearing functional kleptoplasts is less apparent (Rauch et al. 2017). For instance, it is estimated that kleptoplasts can provide up to 60% of the required carbon (Raven et al. 2001; Rauch et al. 2017), while symbionts in other photosymbioses can fully support the host up to over 170% carbon (Thomas et al. 2023). To understand the nutritional relevance of kleptoplasts to sacoglossans, starvation experiments under continuous light (photosynthetic condition) and constant darkness (non-photosynthetic condition) are often conducted. Several studies have shown that dark-starved animals lose body size and weight faster than light-starved animals (Cartaxana et al. 2017; Shiroyama et al. 2020). In others, the effects of starvation in darkness were only apparent after several weeks (Casalduero and Muniain 2008; Maeda et al. 2021). The differences between dark-starved and light-starved slugs have been mainly interpreted as a result of blocked photosynthetic activity. However, some species can even overcome several months of starvation without functional kleptoplasts under natural light conditions (Klochkova et al. 2013). Further, alternative explanations are often not considered (Christa et al. 2014b). For instance, continuous darkness is an unnatural condition that can affect the slug's metabolism independently of kleptoplasts’ photosynthesis or the slugs’ behavior. Thus, some studies have used a photosynthesis blocker (e.g., Monolinuron) to avoid darkness-induced artifacts and showed similar starvation tolerances compared to photosynthetic conditions (Christa et al. 2014a; de Vries et al. 2015; Laetz et al. 2017b).
Regardless of the exact support by the kleptoplasts, there is no doubt that slugs can metabolize the kleptoplasts’ photosynthates (Kremer 1976; Hinde 1978; Cruz et al. 2020). However, how they access these photosynthates is unclear. From corals, it is known that the host supports the photobionts actively with bicarbonate and ammonium, while the photobionts provide sugars to the host (Barott et al. 2015). This exchange of nutrients is crucial for a successful and long-term symbiosis and requires a symbiosome (Mohamed et al. 2016; Matthews et al. 2017). The symbiosome is a membrane that surrounds the symbiont and contains transporters relevant for translocating ammonium and bicarbonate to the symbiont and glucose to the host (Thies et al. 2022). The disruption of this nutrient exchange may lead to the breakdown of the symbiosis (Rädecker et al. 2021). In sacoglossan sea slugs, it is unknown whether there are nutrient transporters in the membrane surrounding the kleptoplasts and how sacoglossan sea slugs can access the photosynthetically derived assimilates. In some species, kleptoplasts accumulate starch (Evertsen and Johnsen 2009; Pelletreau et al. 2014; Laetz et al. 2017b; Cruz et al. 2020), which at least questions an active export of glucose. It even appears that the slugs digest the kleptoplasts while feeding (Maeda et al. 2012; Frankenbach et al. 2021) and during starvation (Laetz et al. 2017a) to access the photosynthates. Hence, the kleptoplasts may instead function as larders and their products as reserves (Pelletreau et al. 2014; Laetz et al. 2017b) accessed by digestion.
In the present study, we analyzed the gene expression profile of the plastid-bearing sea slug Elysia viridis (Montagu, 1804) under the presence and absence of light to address the effects of darkness on starvation specifically. We focused on energy metabolism, mitochondrial function, and transporters relevant to the exchange of nutrients between the host and the symbiont to maintain symbiosis. We further combined the gene expression of central components of autophagy with the analyses of kleptoplast and lysosome abundance to better understand how the slugs might access the kleptoplasts' photosynthates.
Material and methods
Sea slug collection and laboratory culturing
The specimens of Elysia viridis used for transcriptome analyses were collected from their food source Codium tomentosum Stackhouse, 1797 in September 2020 at the rocky beach of Figueira da Foz, Portugal (40°10′05.0"N, 8°53′20.9"W) and immediately transported to the laboratory. Animals were kept in sets of 10 individuals in 500 ml of natural GF/F (0.7 µm pore size, Whatman) filtered seawater at 18 ± 1 °C, 25–30 µmol photons m−2 s−1 (white fluorescence lamps, Philips, TL-D 36W/54) at a 12 h/12 h day/night cycle and a salinity of 33 PPT. We provided the macroalgae Bryopsis hypnoides J.V.Lamouroux, 1809 as a food source as it can be easily cultured under laboratory settings and it is a source of chloroplasts that remain functional in E. viridis (Rauch et al. 2018). Bryopsis hypnoides was cultured in autoclaved seawater enriched with F/2 Medium (Guillard 1975) under constant aeration and the same light regime and temperature as the slugs. Weekly, 80% of the seawater and 100% medium were changed. The slugs remained in culture five months before the experiment to ensure they were fully acclimated to laboratory conditions.
Experimental setup, RNA extraction, and sequencing
Nine randomly chosen slugs (BioSample IDs: SAMN26303207—SAMN26303215) were used in a starvation experiment. Each slug was transferred into a 50 ml Falcon tube filled with 35 ml natural GF/F filtered seawater, with 50% of the seawater changed daily. After an acclimation period of three weeks, the slugs were randomly chosen to starve in constant darkness for one week (SD; n = 3), to starve for one week under culturing light conditions (SL; n = 3) or to serve as the fed control group under culturing light conditions (FED; n = 3). All slugs (SD, SL, and FED) were fixed in liquid nitrogen at the end of the experiment. All animals were subsequently freeze-dried at −80 °C for two days. Total RNA from each sample was extracted using the my-Budget RNA Mini Kit (Bio-Budget Technologies, Germany) following the manufacturer’s instructions. The RNA concentration was quantified with Qubit fluorometric quantification (ThermoFisher, USA) and NanoDrop™ One/OneC (ThermoFisher, USA). Poly(A) mRNA enrichment, library preparation using NEBNext® Ultra™ RNA Library Prep Kit for Illumina® (New England Biolabs, USA), and 150 bp paired-end sequencing using the Illumina NovaSeq system was done by Novogene (UK).
Transcriptome assembly and annotation
The sequencing resulted in 709,603,664 quality filtered (Q30) paired-end reads (on average 78,844,851 per library; Online Resource 1; BioProject ID: PRJNA810987, Accession Nr.: SRR18163429 – SRR18163437). Reads were assembled using Trinity v2.12.0 (Grabherr et al. 2011), and the assembled transcriptome was then clustered with CD-HIT-EST v.4.8.1 with -c 0.9 -n 7 -B 1 -g 1 -s 0.9 parameters (Fu et al. 2012) (Online Resource 2). For each contig, the longest open reading frames were obtained using TransDecoder v5.5.0 (Haas and Papanicolaou 2015). The data set was then annotated by a BLASTP search (part of the BLAST + package v2.9.0) against the UniProt database version 09/2021 (The UniProt Consortium 2019). Subsequently, all non-annotated genes were subjected to a BLASTP search against a molluscan-specific TrEMBL database. Taxonomic assignment for each protein sequence was obtained from the UniProt and TrEMBL taxonomic database (Online Resource 3). Sequences and annotations were subsequently filtered for metazoan annotations to exclude annotations of bacteria, fungi, and plants in downstream analyses. Functional annotations of gene ontology (GO) terms, cluster of Eukaryotic Orthologous groups (KOG), and Kyoto Encyclopedia of Genes and Genomes (KEGG) identifiers for the filtered data set were obtained by using eggNOG mapper v.2.1.6 in default mode (Huerta-Cepas et al. 2017) (Online Resource 4).
Differential gene expression analysis
The transcript abundance of sequences was estimated with kallisto v0.46.2 (Bray et al. 2016) following the Trinity pipeline v2.12.0 (Online Resource 1) to obtain raw read counts and normalized trimmed mean of M-values (TMM). Differential expression analysis was performed with DeSeq2 v1.32.0 (Love et al. 2014) using the raw reads matrix implemented in Trinity. Differentially expressed genes (DEGs) were defined by a false discovery rate (FDR) < 0.05. Principal Component Analysis (PCA) of annotated DEGs was done using the prcomp() function in the stats package in R v. 4.1.0 (R Core Team 2020) using the TMM values for each gene to visualize the effects of starvation on the general gene expression. Gene ontology analyses with adaptive clustering were performed using the GOMWU R package to measure whether each GO category was significantly enriched according to the expression value (Wright et al. 2015). EuKaryotic Orthologous Groups (KOG) class enrichment tests were performed using the KOGMWU R Package (Dixon et al. 2015). KOGMWU allows the comparison of the gene expression of samples analyzed under different conditions. The correlation is based on the KOG delta ranks across the data sets. The delta rank is the difference between the mean rank of genes in a KOG class and the mean rank of all other genes (Dixon et al. 2015). To get a general idea of how starvation under different light conditions affects E. viridis, we compared our data set with published data of developing juveniles of Elysia chlorotica (Chan et al. 2018), heat-stressed colonies of Stylophora pistillata (Esper, 1792) (Rädecker et al. 2021), and symbiotic vs. apo-symbiotic Exaiptasia diaphana (Rapp, 1829) (Lehnert et al. 2014).
DNA extraction, sequencing, and mitochondrial genome assembly and annotation
High molecular weight DNA was extracted using the approach described in Melo Clavijo et al. (2021) from one specimen of E. viridis collected in Aguda, Portugal (41°02′52.6"N, 8°39′16.0"W) in September 2017. The sample was fixed in 70% EtOH and was transported to the University of Aveiro. Single-end library prep of 500–600 bp read length for Illumina HiSeq2500 instrument (1 × 100 nucleotides) was prepared using the tagmentation protocol (Picelli et al. 2014). Overall, 54,437,385 raw single-end Illumina reads with lengths between 50 and 95 nucleotides [nt] (SRR18210511 and SRR18210512) were quality trimmed using BBDuk v.38.91 of the BBMap tool (Bushnell et al. 2017) setting trimq = 20. Sequence quality was subsequently verified using fastqc v.0.11.9 (Andrews 2010). We then used a hybrid assembly approach using Spades v.3.11.1 in default mode (Antipov et al. 2016), including 1,159,884 Nanopore long reads (SRR18251374) generated using the LSK109 kit following the protocol in Melo Clavijo et al. (2021). The mitochondrial genome was identified using a BLASTN search of a cytochrome oxidase 1 nucleotide sequence of Elysia viridis (GenBank accession: MN223463) against the assembly. The extracted contig had a length of 14,204 base pairs (bp) and was subsequently manually edited to obtain the circular genome. The circular sequence consisted of 14,149 bp and was annotated using the MITOS2 webserver (Donath et al. 2019). The annotation was manually curated using Geneious Prime v.2022.0.2 (https://help.geneious.com/hc/en-us/articles/360044627352-How-do-I-cite-Geneious-or-Geneious-Prime-in-a-paper-), using the published mitochondrial genomes of four other Elysia species as reference: E. chlorotica (NC_010567; (Rumpho et al. 2008)), E. cornigera (NC_035489; (Rauch et al. 2017)), E. timida (NC_035490; (Rauch et al. 2017)), and E. ornata (NC_030537; (Karagozlu et al. 2016)). A circular view of the mitochondrial genome was created using OrganellarGenomeDraw (Lohse et al. 2013).
Kleptoplast and lysosomal abundance measurements
Twenty specimens of E. viridis used for kleptoplast and lysosomal abundance analyses were collected in May 2017 in Aguda, Portugal (41°02'46.7″N, 8°39'14.6″W), in rocky intertidal pools and transferred to the laboratory at the Zoological Research Museum Alexander Koenig (Bonn, Germany). Specimens were cultured in groups of 10 individuals in 500 mL artificial seawater (ABReef Salt, Aqua Medic, Germany, ASW) in glass jars with a salinity of 33 PPT under a 12 h/12 h day/night cycle setting the light intensity to 30 µmol photons m−2 s−1 provided by daylight sunrise LED X-change tubes (Sera, Germany) at 20 ± 1 °C. The water was changed three times a week. The food source, Bryopsis hypnoides, was given to the slugs three times a week for five weeks. B.ryopsis hypnoides was cultured under the conditions mentioned above. For each light treatment (SD, SL, and FED), three randomly chosen slugs were used and cultured under the same conditions in individual 50 mL plastic containers filled with 35 mL of ASW. Slugs were imaged using a DP21 camera mounted on an SZX12 stereo microscope (Olympus, Tokyo, Japan) at the beginning of the experiment and, in the case of the SD and SL group, after one week of starvation, and the length measured as an indicator of fitness. The average length of each slug was calculated from five images taken during movement to obtain the point of maximum elongation. Chlorophyll a fluorescence was subsequently measured in each slug using a Diving Pulse Amplitude Modulated (Diving-PAM) fluorometer (Walz, Effeltrich Germany). For this, samples were dark acclimated for 5 min, and then the maximum quantum yield (Fv/Fm; Fv = Fm −Fo; where Fo and Fm are the minimum and maximum fluorescence emitted by dark-adapted samples, respectively) was determined by applying a saturation pulse (pulse duration 0.8 ms, white light, > 5,000 µmol photons m−2 s−1) (Serôdio et al. 2010). One measurement per specimen was performed by placing the optic fiber about 3–5 mm above the slug’s pericardium to obtain F0 values of around 200–500. Measurements were taken at the beginning of the experiment and immediately after one week of starvation before preparing the samples for confocal microscopy and served as a measure of kleptoplast fitness.
To determine the lysosomal and kleptoplast abundance, SD, SL, and FED animals were examined by confocal laser scanning microscopy using a Leica SPE CLSM (Leica Microsystems AG, Germany). Kleptoplasts were detected via chlorophyll a autofluorescence. The lysosome abundance of the slugs was visualized by incubating them for 30 min in 5 µM acridine orange (Sigma-Aldrich, Germany). After incubation, the slugs were vivisected into ~ 0.5 mm thick transverse sections. The samples were then placed on a microscope slide and imaged using a 488 nm laser to excite both acridine orange and chlorophyll a. The wavelength range accepted for acridine orange emission was set to 645 nm to 770 nm to distinguish acridine orange from chlorophyll a (acridine orange dimer peak emission is 656 nm) (Laetz et al. 2017a). Chlorophyll a autofluorescence was recorded, setting the emission from 600–640 nm (peak 633 nm). Five different sections per slug were imaged. Each section was made up of 8 images 1 μm in depth (z plane) for a total of 8 μm of tissue depth. Thus, for each experimental condition (SD, SL, FED), 15 image stacks were analyzed. The images were processed in ImageJ v. 1.52i (Schindelin et al. 2012) by defining a region of interest and subsequently quantifying the fluorescence signals. All images were processed using the same settings for the fluorescence signals.
Results
Starvation, particularly in darkness, induced changes in energy metabolism
The transcriptome of Elysia viridis consisted of 366,188 contigs that translated into 104,180 amino acid sequences based on potential open reading frames. We could annotate 22,996 (22.07% of all protein sequences) of these amino acid sequences, most likely belonging to E. viridis (Online Resource 5). Additionally, 205 protein sequences (0.20%) could be assigned to Archaea, 10,599 sequences (10.17%) were assigned to Bacteria, and 925 sequences (0.89%) were assigned to Viridiplantae (Online Resource 3). For all subsequent analyses, we only used the genes that most likely belonged to E. viridis.
We analyzed the gene expression in Elysia viridis and compared freshly fed (FED) animals and those starved for one week under a 12 h / 12 h day/night light cycle (SL) or in complete darkness (SD). Generally, SD resulted in a different gene expression than SL; both were different from the gene expression of FED specimens. An exception was a replicate of SL animals (SL02) that showed little correlation with the other samples (Fig. 1A) and showed a higher PC variance compared to the additional replicates (Fig. 1B). Thus, this sample was excluded from the subsequent differential gene expression analyses. Overall, we identified 1,862 differentially expressed genes (DEGs) when comparing FED and SD samples, 216 DEGs when comparing FED and SL samples, and 879 DEGs when comparing SL and SD samples (Online Resource 5).
We found no significantly enriched Gene Ontology (GO) term among DEGs when using GOMWU to compare FED and SL animals (Online Resource 6). In the Biological Process (BP) division, we found 22 significantly enriched GO-terms (FDR < 0.1) when comparing SD to FED animals and 24 significantly enriched GO-terms (FDR < 0.1) when comparing SD to SL animals (Fig. 2). The majority of these GO terms were down-regulated in SD animals and could be assigned to the cell cycle and development, independent of the comparison (Fig. 2). Two GO terms related to translation and the GO term "cellular amino acid metabolism" were enriched and up-regulated in SD animals. The GO term "protein folding" was up-regulated when comparing SD to SL animals. GO terms associated with the "cytoskeleton", "microtubule organization center", and "cilium" in the cellular compartment (CC) division were enriched and down-regulated in SD animals, independent of the comparison; The GO term "ribosome" was enriched and up-regulated (Online Resource 6). In the Molecular Function (MF) division, the GO terms "cytoskeletal protein binding" and "transcription factor binding" were enriched and down-regulated in SD compared to FED animals. In contrast, the GO terms "ligase activity", "lyase activity", and "isomerase activity" were enriched and up-regulated (Online Resource 6).
Gene ontology (GO) terms significantly enriched in the Biological Process (BP) category with genes either up- (red) or down-regulated (blue) in specimens of Elysia viridis starved in darkness (SD) compared to freshly fed (FED) or light starved (SL) animals. Shown are genes with an FDR < 0.05 relative to the total number of all genes within the BP category
We analyzed the euKaryotic Orthologous Groups (KOG) using KOGMWU to investigate further the general expression profile (Online Resource 7). We found one main cluster made of fed individuals of Elysia chlorotica and one main cluster made of our starved samples of Elysia viridis and two selected cnidarians (Fig. 3). The expression of genes assigned to the respective KOG categories of SD animals, independent of the comparison with FED or SL animals, clustered with individuals of Exaiptasia diaphana (comparison of symbiotic vs. apo-symbiotic animals). The gene expression of SL vs. FED individuals of E. viridis clustered with heat-stressed individuals of Stylophora pistillata (Fig. 3).
Heat map of enrichment of cluster of Eukaryotic Orthologous groups (KOG) classes by differentially expressed genes in different data sets. Significantly enriched KOG classes (FDR < 0.05) with up- (red) or down-regulated (blue) genes are identified by an asterisk. For data references, see the text. Different letters after Elysia chlorotica indicate stable symbiosis (S), transient symbiosis (U), and stable symbiosis (T), as defined earlier (Pelletreau et al. 2012)
Similarly, as in the Gene Ontology analyses, we could not find significantly enriched KOG categories in SL vs. FED individuals of E. viridis (Fig. 3). In SD animals, the categories "Energy production and conversion" and "Cell wall/membrane/envelope biogenesis" were enriched with mainly up-regulated genes, regardless of being compared to FED or SL animals. Moreover, in SD vs. SL animals, the KOG categories "Nucleotide transport and metabolism", "Amino acid transport and metabolism", and "Carbohydrate transport and metabolism" were enriched among up-regulated genes in SD animals (Fig. 3).
The KOG categories "RNA processing and modification", "Cytoskeleton", and "Signal transduction mechanisms" were enriched and down-regulated independent of the comparison of SD animals, supporting our GO enrichment analyses. Furthermore, the KOG categories "Intracellular trafficking, secretion, and vesicular transport" and "Posttranslational modification, protein turnover, chaperones" were enriched and down-regulated in the SD vs. FED comparison (Fig. 3).
Protein import into mitochondria decreases during starvation
Many enriched and down-regulated GO and KOG terms were assigned to energy metabolism. Thus, we subsequently analyzed the gene expression (not only of the differentially expressed genes) associated with the mitochondria in more detail. In starved specimens, independent of the light condition, the oxidative phosphorylation (OXPHOS) was predominantly down-regulated, especially genes belonging to complex I, II, and V (Online Resource 5). In SD animals, various genes of the Pentose Phosphate Pathway (e.g., deoxyribose-phosphate aldolase), the tricarboxylic acid cycle (e.g., oxogluterate dehydrogenase), and glycolysis (e.g., 6-phosphofructinase, pyruvate dehydrogenase) were up-regulated (Fig. 4A; Online Resource 5). Most genes belonging to the fatty acid metabolism were down-regulated in starved animals (Fig. 4A), including carnitine O-palmitoyltransferase I and II (CPT1 and CPT2). However, several genes involved in fatty acid beta-oxidation in SD animals were up-regulated (Fig. 4B).
A Overview of the expression of mitochondrial energy-related genes in E. viridis freshly fed (FED), starved in darkness (SD), and starved in light (SL). The average TMM of the replicates of each condition in relation to the average TMM of the respective gene across all samples is shown. B Specific expression profile of important genes related to fatty acid metabolism (purple), Pentose Phosphate Pathway (yellow), genes of the Translocase of the inner (TIM, pink) and outer (TOM, orange) mitochondrial membrane. Boxplot whiskers show the minimum and maximum TMM values, and the centers correspond to the medians. C Circular map of the mitochondrial genome of Elysia viridis. The annotations of the tRNAs were omitted for displaying reasons. ACAD: acyl-CoA dehydrogenase; ACAT: acetyl-CoA acetyltransferase; ACS: acetyl-CoA synthetase; CPT1, CPT2: Carnitine palmitoyltransferase 1, 2; ECHD: enoyl-CoA hydratase; HADH: hydroxyacyl-CoA dehydrogenase; DERA: deoxyribose-phosphate aldolase; TMM: Trimmed means of M value; TCA: tricarboxylic acid cycle; OXPHOS: oxidative phosphorylation; PPP: pentose phosphate pathway
The mitochondrial genome of E. viridis (GenBank accession no.: ON065001) has a length of 14,149 bp and consists of 13 protein-coding genes, 2 rRNAs, and 21 tRNAs, a standard set of genes among Heterobranchia, except for a missing tRNA-S2 gene. A comparison with the published mitochondrial genomes of four other Elysia species did not reveal sufficient sequence/structure conservation to annotate this gene. Most proteins relevant to mitochondrial metabolism are nuclear-encoded and have to be imported. Yet, core components of the translocase of the outer membrane (TOM) and the translocase of the inner membrane (TIM) complexes were down-regulated in starved slugs (Fig. 4B).
Autophagy as primary source to obtain kleptoplast nutrients
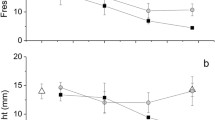
Afterwards, we examined how starving slugs may obtain nutrients by analyzing the abundance of lysosomes and the expression of genes involved in lysosome formation and autophagy. Independent of the light condition, starvation for one week led to an increase in the abundance of lysosomes and a simultaneous decrease in the abundance of kleptoplasts in specimens of E. viridis (Fig. 5A–C). SD animals had a higher lysosomal abundance (39.9% ± 5.7%) after one week of starvation than SL animals (17.3% ± 1.7%) and FED animals (11.4% ± 2.1%; Fig. 5B). There was a substantial decrease in kleptoplasts abundance during starvation compared to FED animals (42.9% ± 5.1%; Fig. 5B). Kleptoplast abundance in the starvation conditions was low, albeit a considerable variation in SD animals was observed (SD: 15.7% ± 7.9%, SL 19.7% ± 1.7%; Fig. 5C). Interestingly, SL animals slightly increased in length after one week of starvation (102% ± 4% of the initial length), while in SD animals, the length was reduced by about 3% (97.0% ± 2.0% of the initial length; Fig. 5D). The maximum quantum yield (Fv/Fm) of kleptoplasts decreased similarly in SD and SL specimens (Fig. 5E).
A Confocal images of the digestive gland tubules of a lateral part of the parapodia and microscopic images of single individuals of E. viridis freshly fed (FED), starved for one week in continuous darkness (SD), and starved for one week in light (SL). The auto-fluorescence of chlorophyll a of kleptoplasts (red) and the fluorescence of acridine orange depicting lysosomal abundance (blue) are shown. Percentage of lysosomes (B) and kleptoplasts (C) coverage in the digestive gland tubules (DGS) of 5 different regions in the parapodia of individuals of E. viridis freshly fed (FED) and after one week of starvation in continuous darkness (SD) or light (SL). D Relative length of individuals of E. viridis during one week of starvation in continuous darkness (SD) or light (SL), and (E) the maximum quantum yield (Fv/Fm) of the kleptoplasts in the same individuals. Boxplot whiskers show the minimum and maximum values, and the center lines correspond to the medians
Consistent with an increased lysosomal abundance, the serine/threonine-protein kinase TOR (target of rapamycin) was down-regulated in starved animals, which may initiate autophagy. Beclin-1 (BECN) was highly up-regulated in SD animals but only slightly in SL animals. Additionally, VPS34 was down-regulated in SD slugs, which can also trigger autophagy (Fig. 6A). Several lysosomal cathepsins (CTSB, CTSD, and CTSF), serine carboxypeptidase (CPVL), and NPC intracellular cholesterol transporter 2 (NPC2) were up-regulated during starvation, independent of the light conditions (Fig. 6A). On the contrary, lysosomal genes like alpha-glucosidase (GAA) and alpha-mannosidase (MANB) were down-regulated in starved animals (Fig. 6A).
A Simplified pathway and (B) expression of genes involved in autophagy-related components and downstream formation of autophagolysosomes of E. viridis freshly fed (FED) during one week of starvation in continuous darkness (SD) or the light (SL). The pathway was created with BioRender.com. mt: mitochodria; cp: chloroplasts; lp: lipids; pr: proteins. C Expression of genes of the GS-GOGAT cycle of E. viridis freshly fed (FED) during one week of starvation in continuous darkness (SD) or the light (SL). Boxplots show the expression values in TMM (Trimmed means of M values), where the whiskers are the minimum and maximum values, and the centers correspond to the medians. mTOR: mechanistic target of rapamycin kinase; ATG101: autophagy related 101; BECN: beclin; VPS34: also PIK3C3: phosphatidylinositol 3-kinase catalytic subunit type 3; CTSB, CTSD, CTSF: cathepsin B, D, F; CPVL: carboxypeptidase vitellogenic like; GAA: alpha glucosidase; MANB: phosphomannomutase; NPC2: NPC intracellular cholesterol transporter 2; GS: glutamine synthetase; GOGAT: glutamate synthase; GDH ana.: glutamate dehydrogenase anabolic; GDH cat: glutamate dehydrogenase catabolic
Glutamine is generated from ammonia by glutamine synthetase (GS), which was up-regulated in SL and SD slugs (Fig. 6B). The glutamate synthase (GOGAT) and the catabolic glutamate dehydrogenase (GLUD2) were also up-regulated, especially in SD animals, while the anabolic glutamate dehydrogenase (GLUD1) was primarily down-regulated in SL animals (Fig. 6B).
There was no up-regulation of bicarbonate, carbonic anhydrases, or glucose transporters regardless of the light condition, but only a significant up-regulation of ammonium transporters in SD animals (Online Resource 5).
Discussion
Even after decades of research, several aspects of functional kleptoplasty in Sacoglossa remain unclear. Of particular interest is the contribution of kleptoplasts to the metabolism of the slugs and how the slugs can access the photosynthates of the kleptoplasts. Continuous darkness is often used to assess the contribution of photosynthetically active kleptoplasts to the slug’s metabolism (Casalduero and Muniain 2008; Yamamoto et al. 2013; Christa et al. 2014a; Cartaxana et al. 2017). However, it remains unexplored if and how darkness affects the metabolism of sacoglossan sea slugs. This is the first study to compare the gene expression of a plastid-bearing sea slug starved in light and continuous darkness to shed light on the effects of starvation in darkness. We found that darkness primarily leads to glucose starvation and a general down-regulation of the metabolism. However, in conjunction with an increased autophagy rate regardless of the light condition, our results support the hypothesis that kleptoplasts function as a sort of reserve or larder in Elysia viridis.
Dark-starved slugs reduce their metabolic activity, cellular processes, and development. Further, starvation in darkness leads to a metabolic switch towards fatty acid beta-oxidation and deoxyribose-phosphate aldolase (DERA) expression. Interestingly, both are induced by glucose starvation in organisms (Salleron et al. 2014; Weber et al. 2020), which is observed in dark-starved specimens and could be related to a lack of glucose produced through photosynthesis. Starvation also caused the down-regulation of the translocase complexes TIM and TOM and fatty acid translocase receptors (CPTI and CPTII) of mitochondria. This down-regulation reduces the capacity to import nuclear-encoded proteins and translocate fatty acid into the mitochondria. Particularly during periods without food, CPTII deficiency prevents using certain fats for energy production (Joshi and Zierz 2020). Hence, starvation in darkness seems to induce a metabolic switch towards fatty acid degradation, but energy production through this pathway is highly reduced and independent of the light condition. Despite this, starvation in darkness might not only induce glucose starvation. Among others, constant darkness could inhibit the slug's diurnal rhythm, leading to a stress response similar to that observed in other gastropods (Shirley and Findley 1978). Darkness could also induce a resting phase, irrespective of food availability. Our KOG analyses further suggest a comparable stress response in different taxa to diverse factors like starvation, higher temperature, or darkness. Hence, at this stage, an unambiguous assignment of what triggers the reduction in the metabolic and developmental processes in dark-starved slugs is complicated. Nevertheless, the absence of photosynthates, specifically glucose, may serve as a starvation enhancer, if not an inducer. Thus, kleptoplast photosynthesis can avoid metabolic reduction by providing glucose, at least for one week of starvation in Elysia viridis.
It has been shown that kleptoplasts accumulate starch in some species (Evertsen and Johnsen 2009; Laetz et al. 2017b). During feeding, the kleptoplasts are exchanged (Maeda et al. 2012; Melo Clavijo et al. 2021), and an increased lysosomal abundance during starvation points towards digestion of the kleptoplasts (Laetz et al. 2017a). Therefore, kleptoplasts may export a few photosynthates to slugs actively or not at all. The increase in lysosomal abundance and decrease in chloroplasts abundance that we observed here, provide further support that E. viridis digests its chloroplasts to acquire energy when autophagy is initiated.
Autophagy is probably the primary energy source during starvation, irrespective of starvation in light or darkness, as previously suggested (de Vries et al. 2014), but there are notable differences between starvation in light and darkness. The increased lysosomal abundance in darkness compared to light is likely based on a lower nutritional value of the kleptoplasts in non-photosynthetic conditions. Hence, more slug tissue is needed to satisfy the energetic demands. In light, kleptoplasts accumulate photosynthates (Laetz et al. 2017a) and provide sufficient amounts of glucose (or starch) to prevent physiological responses to glucose starvation and increased need for slug tissue degradation. This could also explain the slight increase in the length of the slugs in light and the decrease in the length of slugs in darkness. Similar observations were made in juveniles of Elysia chlorotica in which a slight increase in length was observed shortly after entering starvation, followed by a decrease in length in the longer term (Pelletreau et al. 2012). As long as the kleptoplasts are fully functional, as suggested by our Chl a fluorescence measurement, they can provide a benefit to the slugs, at least for a couple of days to one week, by acting as some sort of food depot.
Our results suggest that the autophagic response could be initiated via the down-regulation of VPS34 due to glucose starvation (Corona Velazquez and Jackson 2018). Since SL animals might not enter glucose starvation, a different pathway is likely involved, but the exact mechanism needs to be further examined. Moreover, even under feeding conditions, we could not find an expression of transporters relevant for nutrient exchange in other photosymbioses, including glucose, ammonium, and bicarbonate transporters (Zoccola et al. 2015; Matthews et al. 2017; Roberty et al. 2020) further supporting the lack of an active nutrient exchange.
Apart from glucose, ammonium is a crucial nutrient that connects hosts and their symbionts in photosymbioses via the GS/GOGAT cycle. It was recently suggested that functional kleptoplasts under constant feeding and light could putatively help to retrieve additional ammonium via photosynthesis in Elysia viridis (Cruz et al. 2020). Our results revealed an up-regulation of both GS and GOGAT during starvation in E. viridis, indicating that the slugs indeed receive an increased amount of ammonium. The primary source of ammonium under starvation conditions are macromolecules, such as proteins, that are degraded by autophagy (Liu et al. 2021). Hence, starvation for one week probably does not induce a nitrogen limitation, and the kleptoplasts may serve as a nitrogen source.
Furthermore, in other photosymbioses, the translocation of bicarbonate to the symbionts is vital to ensure a high photosynthetic rate (Matthews et al. 2017; Roberty et al. 2020). In some sacoglossans, the maximum quantum yield (Fv/Fm) of kleptoplasts is higher compared to the Fv/Fm of the chloroplasts in their natural host algae (Serôdio et al. 2014). Because we could not identify an up-regulation of relevant bicarbonate transporters, this increased Fv/Fm could be based on a higher partial CO2 pressure inside the animal cells that would induce a higher CO2 availability for the kleptoplasts through diffusion (Serôdio et al. 2014).
Conclusion
Kleptoplasts in sacoglossans synthesize photosynthates when provided light and can act as a valuable source of nutrients during starvation at least for one week. Under non-photosynthetic conditions, the lack of nutritional support induces length reduction of the slug. Here we show that autophagy is probably a primary source of nutrients for the slugs during starvation in light or darkness. Starvation in darkness reduces energy-consuming processes, such as cell cycle and development, which is probably caused by glucose starvation. The photosynthetic activity of kleptoplasts may help to overcome some energetic shortcomings, but photosynthates are probably not actively exported to the slug. Kleptoplasts in E. viridis more likely function as larders, whose energy can be utilized following their degradation during autophagy and can support the slugs under photosynthetic conditions in the short term.
Data availability
All raw reads of the RNA sequencing and the DNA sequencing are available at GenBank under the Bioproject ID PRJNA810987. The RNA assembly is available as Online Resource.
References
Andrews S (2010) FastQC: A quality control tool for high throughput sequence data
Antipov D, Korobeynikov A, McLean JS, Pevzner PA (2016) hybridSPAdes: an algorithm for hybrid assembly of short and long reads. Bioinformatics 32:1009–1015. https://doi.org/10.1093/bioinformatics/btv688
Barott KL, Venn AA, Perez SO, Tambutté S, Tresguerres M (2015) Coral host cells acidify symbiotic algal microenvironment to promote photosynthesis. Proc Natl Acad Sci 112:607–612. https://doi.org/10.1073/pnas.1413483112
Bray NL, Pimentel H, Melsted P, Pachter L (2016) Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol 34:525–527. https://doi.org/10.1038/nbt.3519
Bushnell B, Rood J, Singer E (2017) BBMerge – accurate paired shotgun read merging via overlap. PLoS ONE 12:e0185056. https://doi.org/10.1371/journal.pone.0185056
Cartaxana P, Trampe E, Kühl M, Cruz S (2017) Kleptoplast photosynthesis is nutritionally relevant in the sea slug Elysia viridis. Sci Rep 7:7714. https://doi.org/10.1038/s41598-017-08002-0
Cartaxana P, Morelli L, Quintaneiro C, Calado G, Calado R, Cruz S (2018) Kleptoplasts photoacclimation state modulates the photobehaviour of the solar-powered sea slug Elysia viridis. J Exp Biol jeb. https://doi.org/10.1242/jeb.180463
Cartaxana P, Morelli L, Jesus B, Calado G, Calado R, Cruz S (2019) The photon menace: kleptoplast protection in the photosynthetic sea slug Elysia timida. J Exp Biol 222:jeb202580. https://doi.org/10.1242/jeb.202580
Casalduero FG, Muniain C (2008) The role of kleptoplasts in the survival rates of Elysia timida (Risso, 1818): (Sacoglossa: Opisthobranchia) during periods of food shortage. J Exp Mar Biol Ecol 357:181–187. https://doi.org/10.1016/j.jembe.2008.01.020
Chan CX, Vaysberg P, Price DC, Pelletreau KN, Rumpho ME, Bhattacharya D (2018) Active host response to algal symbionts in the sea slug Elysia chlorotica. Mol Biol Evol 35:1706–1711. https://doi.org/10.1093/molbev/msy061
Christa G, Zimorski V, Woehle C, Tielens AGM, Wägele H, Martin WF, Gould SB (2014a) Plastid-bearing sea slugs fix CO2 in the light but do not require photosynthesis to survive. Proc R Soc B Biol Sci 281:20132493. https://doi.org/10.1098/rspb.2013.2493
Christa G, de Vries J, Jahns P, Gould SB (2014) Switching off photosynthesis: the dark side of sacoglossan slugs. Commun Integr Biol 7:e28029. https://doi.org/10.4161/cib.28029
Christa G, Pütz L, Sickinger C, Melo Clavijo J, Laetz EMJ, Greve C, Serôdio J (2018) Photoprotective Non-photochemical quenching does not prevent kleptoplasts from net photoinactivation. Front Ecol Evol 6:121. https://doi.org/10.3389/fevo.2018.00121
Corona Velazquez AF, Jackson WT (2018) So many roads: the multifaceted regulation of autophagy induction. Mol Cell Biol. https://doi.org/10.1128/MCB.00303-18
Cruz S, LeKieffre C, Cartaxana P, Hubas C, Thiney N, Jakobsen S, Escrig S, Jesus B, Kühl M, Calado R, Meibom A (2020) Functional kleptoplasts intermediate incorporation of carbon and nitrogen in cells of the Sacoglossa sea slug Elysia viridis. Sci Rep 10:10548. https://doi.org/10.1038/s41598-020-66909-7
de Vries J, Christa G, Gould SB (2014) Plastid survival in the cytosol of animal cells. Trends in Plant Sci 19:347–350. https://doi.org/10.1016/j.tplants.2014.03.010
de Vries J, Woehle C, Christa G, Wägele H, Tielens AGM, Jahns P, Gould SB (2015) Comparison of sister species identifies factors underpinning plastid compatibility in green sea slugs. Proc R Soc B Biol Sci 282:20142519. https://doi.org/10.1098/rspb.2014.2519
Dixon GB, Davies SW, Aglyamova GV, Meyer E, Bay LK, Matz MV (2015) Genomic determinants of coral heat tolerance across latitudes. Science 348:1460–1462. https://doi.org/10.1126/science.1261224
Donath A, Jühling F, Al-Arab M, Bernhart SH, Reinhardt F, Stadler PF, Middendorf M, Bernt M (2019) Improved annotation of protein-coding genes boundaries in metazoan mitochondrial genomes. Nucleic Acids Res 47:10543–10552. https://doi.org/10.1093/nar/gkz833
Evertsen J, Johnsen G (2009) In vivo and in vitro differences in chloroplast functionality in the two north Atlantic sacoglossans (Gastropoda, Opisthobranchia) Placida dendritica and Elysia viridis. Mar Biol 156:847–859. https://doi.org/10.1007/s00227-009-1128-y
Frankenbach S, Luppa Q, Serôdio J, Greve C, Bleidissel S, Melo Clavijo J, Laetz EMJ, Preisfeld A, Christa G (2021) Kleptoplasts are continuously digested during feeding in the plastid-bearing sea slug Elysia viridis. J Molluscan Stud 87:eyab022. https://doi.org/10.1093/mollus/eyab022
Fu L, Niu B, Zhu Z, Wu S, Li W (2012) CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28:3150–3152. https://doi.org/10.1093/bioinformatics/bts565
Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, Mauceli E, Hacohen N, Gnirke A, Rhind N, di Palma F, Birren BW, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29:644–652. https://doi.org/10.1038/nbt.1883
Guillard RRL (1975) Culture of Phytoplankton for Feeding Marine Invertebrates. In: Smith WL, Chanley MH (eds) Culture of Marine Invertebrate Animals. Springer, US, Boston, MA, pp 29–60
Haas B, Papanicolaou A (2015) TransDecoder (find coding regions within transcripts). Github, https://github.com/TransDecoder/TransDecoder.
Händeler K, Grzymbowski YP, Krug PJ, Wägele H (2009) Functional chloroplasts in metazoan cells - a unique evolutionary strategy in animal life. Front Zool 6:28. https://doi.org/10.1186/1742-9994-6-28
Havurinne V, Aitokari R, Mattila H, Käpylä V, Tyystjärvi E (2021) Ultraviolet screening by slug tissue and tight packing of plastids protect photosynthetic sea slugs from photoinhibition. Photosynth Res. https://doi.org/10.1101/2021.07.15.452583
Havurinne V, Tyystjärvi E (2020) Photosynthetic sea slugs induce protective changes to the light reactions of the chloroplasts they steal from algae. eLife 9: e57389 https://doi.org/10.7554/eLife.57389
Hinde R (1978) The metabolism of photosynthetically fixed carbon by isolated chloroplasts from Codium fragile (Chlorophyta: Siphonales) and by Elysia viridis (Mollusca: Sacoglossa). Biol J Linn Soc 10:329–342. https://doi.org/10.1111/j.1095-8312.1978.tb00019.x
Huerta-Cepas J, Forslund K, Coelho LP, Szklarczyk D, Jensen LJ, von Mering C, Bork P (2017) Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Mol Biol Evol 34:2115–2122. https://doi.org/10.1093/molbev/msx148
Joshi PR, Zierz S (2020) Muscle carnitine palmitoyltransferase II (CPT II) deficiency: a conceptual approach. Molecules 25:1784. https://doi.org/10.3390/molecules25081784
Karagozlu MZ, Sung J, Lee J, Kwon T, Kim C-B (2016) Complete mitochondrial genome sequences and phylogenetic relationship of Elysia ornata (Swainson, 1840) (Mollusca, Gastropoda, Heterobranchia, Sacoglossa). Mitochondrial DNA Part B 1:230–232. https://doi.org/10.1080/23802359.2016.1155427
Klochkova TA, Han JW, Chah K-H, Kim RW, Kim J-H, Kim KY, Kim GH (2013) Morphology, molecular phylogeny and photosynthetic activity of the sacoglossan mollusc, Elysia nigrocapitata, from Korea. Mar Biol 160:155–168. https://doi.org/10.1007/s00227-012-2074-7
Kremer BP (1976) Photosynthetic carbon metabolism of chloroplasts symbiotic with a marine opisthobranch. Z Für Pflanzenphysiol 77:139–145. https://doi.org/10.1016/S0044-328X(76)80176-6
Laetz EMJ, Rühr PT, Bartolomaeus T, Preisfeld A, Wägele H (2017a) Examining the retention of functional kleptoplasts and digestive activity in sacoglossan sea slugs. Org Divers Evol 17:87–99. https://doi.org/10.1007/s13127-016-0308-0
Laetz EMJ, Moris VC, Moritz L, Haubrich AN, Wägele H (2017b) Photosynthate accumulation in solar-powered sea slugs - starving slugs survive due to accumulated starch reserves. Front Zool 14:4. https://doi.org/10.1186/s12983-016-0186-5
Lehnert EM, Mouchka ME, Burriesci MS, Gallo ND, Schwarz JA, Pringle JR (2014) Extensive differences in gene expression between symbiotic and aposymbiotic cnidarians. G3-, Gen Genet 4:277–295. https://doi.org/10.1534/g3.113.009084
Liu K, Sutter BM, Tu BP (2021) Autophagy sustains glutamate and aspartate synthesis in Saccharomyces cerevisiae during nitrogen starvation. Nat Commun 12:57. https://doi.org/10.1038/s41467-020-20253-6
Lohse M, Drechsel O, Kahlau S, Bock R (2013) OrganellarGenomeDRAW—a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res 41:W575–W581. https://doi.org/10.1093/nar/gkt289
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. https://doi.org/10.1186/s13059-014-0550-8
Maeda T, Hirose E, Chikaraishi Y, Kawato M, Takishita K, Yoshida T, Verbruggen H, Tanaka J, Shimamura S, Takaki Y, Tsuchiya M, Iwai K, Maruyama T (2012) Algivore or Phototroph? Plakobranchus ocellatus (Gastropoda) continuously acquires kleptoplasts and nutrition from multiple algal species in nature. PLoS ONE 7:e42024. https://doi.org/10.1371/journal.pone.0042024
Maeda T, Takahashi S, Yoshida T, Shimamura S, Takaki Y, Nagai Y, Toyoda A, Suzuki Y, Arimoto A, Ishii H, Satoh N, Nishiyama T, Hasebe M, Maruyama T, Minagawa J, Obokata J, Shigenobu S (2021) Chloroplast acquisition without the gene transfer in kleptoplastic sea slugs plakobranchus ocellatus. eLife 10:e60176. https://doi.org/10.7554/eLife.60176
Matthews JL, Crowder CM, Oakley CA, Lutz A, Roessner U, Meyer E, Grossman AR, Weis VM, Davy SK (2017) Optimal nutrient exchange and immune responses operate in partner specificity in the cnidarian-dinoflagellate symbiosis. Proc Natl Acad Sci 114:13194–13199. https://doi.org/10.1073/pnas.1710733114
Melo Clavijo J, Donath A, Serôdio J, Christa G (2018) Polymorphic adaptations in metazoans to establish and maintain photosymbioses. Biol Rev 93:2006–2020. https://doi.org/10.1111/brv.12430
Melo Clavijo J, Frankenbach S, Fidalgo C, Serôdio J, Donath A, Preisfeld A, Christa G (2020) Identification of scavenger receptors and thrombospondin-type-1 repeat proteins potentially relevant for plastid recognition in Sacoglossa. Ecol Evol. https://doi.org/10.1002/ece3.6865
Melo Clavijo J, Drews F, Pirritano M, Simon M, Salhab A, Donath A, Frankenbach S, Serôdio J, Bleidißel S, Preisfeld A, Christa G (2021) The complete mitochondrial genome of the photosymbiotic sea slug Berghia stephanieae (Valdés, 2005) (Gastropoda, Nudibranchia). Mitochondrial DNA Part B 6:2281–2284. https://doi.org/10.1080/23802359.2021.1914211
Mendoza M, Rocha S, Troncoso J, Posada D, Canchaya CA (2022) Transcriptome landscape of kleptoplastic sea slug Elysia viridis. J Moll Stud 89:001. https://doi.org/10.1093/mollus/eyad001
Mohamed AR, Cumbo V, Harii S, Shinzato C, Chan CX, Ragan MA, Bourne DG, Willis BL, Ball EE, Satoh N, Miller DJ (2016) The transcriptomic response of the coral Acropora digitifera to a competent Symbiodinium strain: the symbiosome as an arrested early phagosome. Mol Ecol 25:3127–3141. https://doi.org/10.1111/mec.13659
Pelletreau KN, Worful JM, Sarver KE, Rumpho ME (2012) Laboratory culturing of Elysia chlorotica reveals a shift from transient to permanent kleptoplasty. Symbiosis 58:221–232. https://doi.org/10.1007/s13199-012-0192-0
Pelletreau KN, Weber APM, Weber KL, Rumpho ME (2014) Lipid accumulation during the establishment of kleptoplasty in Elysia chlorotica. PLoS ONE 9:e97477. https://doi.org/10.1371/journal.pone.0097477
Picelli S, Björklund ÅK, Reinius B, Sagasser S, Winberg G, Sandberg R (2014) Tn5 transposase and tagmentation procedures for massively scaled sequencing projects. Genome Res 24:2033–2040. https://doi.org/10.1101/gr.177881.114
R Core Team (2020) R: A language and environment for statistical computing.
Rädecker N, Pogoreutz C, Gegner HM, Cárdenas A, Roth F, Bougoure J, Guagliardo P, Wild C, Pernice M, Raina J-B, Meibom A, Voolstra CR (2021) Heat stress destabilizes symbiotic nutrient cycling in corals. Proc Natl Acad Sci 118:e2022653118. https://doi.org/10.1073/pnas.2022653118
Rauch C, Jahns P, Tielens AGM, Gould SB, Martin WF (2017) On being the right size as an animal with plastids. Front Plant Sci 8:1402. https://doi.org/10.3389/fpls.2017.01402
Rauch C, Tielens AGM, Serôdio J, Gould SB, Christa G (2018) The ability to incorporate functional plastids by the sea slug Elysia viridis is governed by its food source. Mar Biol 165:82. https://doi.org/10.1007/s00227-018-3329-8
Raven JA, Walker DI, Jensen KR, Handley LL, Scrimgeour CM, McInroy SG (2001) What fraction of the organic carbon in sacoglossans is obtained from photosynthesis by kleptoplastids? An investigation using the natural abundance of stable carbon isotopes. Mar Biol 138:537–545. https://doi.org/10.1007/s002270000488
Roberty S, Béraud E, Grover R, Ferrier-Pagès C (2020) Coral productivity is co-limited by bicarbonate and ammonium availability. Microorganisms 8:640. https://doi.org/10.3390/microorganisms8050640
Rumpho ME, Worful JM, Lee J, Kannan K, Tyler MS, Bhattacharya D, Moustafa A, Manhart JR (2008) Horizontal gene transfer of the algal nuclear gene psbO to the photosynthetic sea slug Elysia chlorotica. Proc Natl Acad Sci USA 105:17867–17871. https://doi.org/10.1073/pnas.0804968105
Salleron L, Magistrelli G, Mary C, Fischer N, Bairoch A, Lane L (2014) DERA is the human deoxyribose phosphate aldolase and is involved in stress response. Biochim Biophys Acta (BBA) - Mol Cell Res 1843:2913–2925. https://doi.org/10.1016/j.bbamcr.2014.09.007
Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez J-Y, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A (2012) Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. https://doi.org/10.1038/nmeth.2019
Schmitt V, Wägele H (2011) Behavioral adaptations in relation to long-term retention of endosymbiotic chloroplasts in the sea slug Elysia timida (Opisthobranchia, Sacoglossa). Thalassas 27:225–238
Serôdio J, Pereira S, Furtado J, Silva R, Coelho H, Calado R (2010) In vivo quantification of kleptoplastic chlorophyll a content in the “solar-powered” sea slug Elysia viridis using optical methods: spectral reflectance analysis and PAM fluorometry. Photochem Photobiol Sci 9:68–77. https://doi.org/10.1039/b9pp00058e
Serôdio J, Cruz S, Cartaxana P, Calado R (2014) Photophysiology of kleptoplasts: photosynthetic use of light by chloroplasts living in animal cells. Philos Trans R Soc B Biol Sci 369:20130242. https://doi.org/10.1098/rstb.2013.0242
Shirley TC, Findley AM (1978) Circadian rhythm of oxygen consumption in the marsh periwinkle, Littorina irrorata (Say, 1822). Comp Biochem Physiol A Physiol 59:339–342. https://doi.org/10.1016/0300-9629(78)90173-1
Shiroyama H, Mitoh S, Ida TY, Yusa Y (2020) Adaptive significance of light and food for a kleptoplastic sea slug: implications for photosynthesis. Oecologia 194:455–463. https://doi.org/10.1007/s00442-020-04779-6
Stanley GD, Lipps JH (2011) Photosymbiosis: The driving force for reef success and failure. Paleontol Soc Pap 17:33–59. https://doi.org/10.1017/S1089332600002436
The UniProt Consortium (2019) UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res 47:D506–D515. https://doi.org/10.1093/nar/gky1049
Thies AB, Quijada-Rodriguez AR, Zhouyao H, Weihrauch D, Tresguerres M (2022) A Rhesus channel in the coral symbiosome membrane suggests a novel mechanism to regulate NH3 and CO2 delivery to algal symbionts. Sci Adv 8:eabm0303. https://doi.org/10.1126/sciadv.abm0303
Thomas NJ, Coates CJ, Tang KW (2023) Environmental constraints on the photosynthetic rate of the marine flatworm Symsagittifera roscoffensis. J Exp Mar Biol Ecol 558:151830. https://doi.org/10.1016/j.jembe.2022.151830
Van Steenkiste NWL, Stephenson I, Herranz M, Husnik F, Keeling PJ, Leander BS (2019) A new case of kleptoplasty in animals: Marine flatworms steal functional plastids from diatoms. Sci Adv 5:eaaw4337. https://doi.org/10.1126/sciadv.aaw4337
Wägele H, Martin WF (2014) Endosymbioses in Sacoglossan Seaslugs: Plastid-Bearing Animals that Keep Photosynthetic Organelles Without Borrowing Genes. In: Löffelhardt W (ed) Endosymbiosis. Springer Vienna, Vienna, pp 291–324
Weaver S, Clark KB (1981) Light intensity and color preferences of five ascoglossan (=sacoglossan) molluscs (Gastropoda: Opisthobranchia): A comparison of chloroplast-symbiotic and aposymbiotic species. Mar Behav Physiol 7:297–306. https://doi.org/10.1080/10236248109386991
Weber CA, Sekar K, Tang JH, Warmer P, Sauer U, Weis K (2020) β-Oxidation and autophagy are critical energy providers during acute glucose depletion in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 117:12239–12248. https://doi.org/10.1073/pnas.1913370117
Wright RM, Aglyamova GV, Meyer E, Matz MV (2015) Gene expression associated with white syndromes in a reef building coral. Acropora Hyacinthus BMC Genom 16:371
Yamamoto S, Hirano YM, Hirano YJ, Trowbridge CD, Akimoto A, Sakai A, Yusa Y (2013) Effects of photosynthesis on the survival and weight retention of two kleptoplastic sacoglossan opisthobranchs. J Mar Biol Assoc U K 93:209–215. https://doi.org/10.1017/S0025315412000628
Zoccola D, Ganot P, Bertucci A, Caminiti-Segonds N, Techer N, Voolstra CR, Aranda M, Tambutté E, Allemand D, Casey JR, Tambutté S (2015) Bicarbonate transporters in corals point towards a key step in the evolution of cnidarian calcification. Sci Rep 5:9983. https://doi.org/10.1038/srep09983
Acknowledgements
We want to thank Thomas Bartolomaeus and the Institute for Evolutionary Biology and Ecology staff for allowing us to use their CLSM and lab facilities and Heike Wägele for providing a space to culture our specimens. This work was supported by the EvoSym project (grant no. POCI-01-0145-FEDER-028751), funded by the Fundo Europeu de Desenvolvimento Regional (FEDER) through the “Programa Operacional Competitividade e Internacionalização” and by national funds from the Fundação para a Ciência e a Tecnologia (FCT)/Ministério da Ciência, Tecnologia e Ensino Superior (MCTES) via the Centre for Environmental and Marine Studies, CESAM (UIDP/50017/2020 + UIDB/50017/2020 + LA/P/0094/2020), funded by national funds (OE) through FCT/MCTES.
Funding
Open Access funding enabled and organized by Projekt DEAL. Centro de Estudos Ambientais e Marinhos,Universidade de Aveiro, POCI-01-0145-FEDER-028751, Gregor Christa
Author information
Authors and Affiliations
Contributions
SF, JS, and GC planned and set up the experiments. SF, JM, MB, CG, CS, LP, EL, and GC performed the experiments. GG and CL prepared the sequencing libraries and performed sequencing. SF, JM, MB, AD, CS, LP, EL, and GC analyzed the data and prepared the figures. All authors drafted and edited the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that the research was conducted without any commercial or financial relationships that could be construed as a potential conflict of interest.
Ethical approval
All applicable international, national, and/or institutional guidelines for the care and use of animals were followed.
Additional information
Responsible Editor: C. Eizaguirre.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Frankenbach, S., Melo Clavijo, J., Brück, M. et al. Shedding light on starvation in darkness in the plastid-bearing sea slug Elysia viridis (Montagu, 1804). Mar Biol 170, 89 (2023). https://doi.org/10.1007/s00227-023-04225-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00227-023-04225-0