Abstract
1H NMR spectroscopy was applied to analyse samples of “Swabian–Hall Quality Pork” with protected geographical indication (PGI). To obtain maximum chemical information sample preparation was based on both polar extraction and non-polar extraction. A non-targeted approach was used to analyse the 1H NMR data followed by principal component analysis (PCA), linear discriminant analysis (LDA), and cross-validation (CV) embedded in a Monte Carlo (MC) resampling approach. A total of 275 raw pork samples were collected in the years 2018 to 2021. The correct prediction rate of “Swabian–Hall Quality Pork” was about 92% on average for both models based on either the polar or non-polar metabolites. In addition, 1H NMR data describing the polar and non-polar metabolites were combined in a classification model to improve the prediction accuracy. By performing a mid-level data fusion, a correct prediction rate of 98% was achieved. Furthermore, spectral regions in the NMR spectra of the polar and non-polar metabolites that are relevant for the classification of the pork samples were identified to describe potential chemical marker compounds.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The old German country pig breed “Swabian–Hall swine” was created in 1820 by crossing the native pig breed “German Landrace” with the Chinese “Meishan” pig with the idea of increasing the fat content of the meat. In the 1950s, there was a high demand for Swabian–Hall swine due to the very good feed conversion and the exceptional fertility of the animals as well as the excellent tasting meat, which is due to the high proportion of fat as a flavor carrier [1, 2]. At that time, the pig population in the northern Württemberg (region of the German state Baden-Württemberg) consisted of 90% Swabian–Hall pigs. However, eating habits changed in the 1960s. Leaner meat was preferred, resulting in a rapid decline in both breeding and demand of the Swabian–Hall pigs and, more generally, all high fat breeds [3, 4]. The Swabian–Hall Breeders Association, which is committed to the preservation of the old landrace, was founded in 1986. The Farmer Producer Association of Swabian Hall, founded in 1988, offers pork that is produced according to binding producer guidelines and that exceed normal requirements. Many consumers ask for meat from animal-friendly husbandry, peasant agriculture, and a regional value chain [1]. The founding motive and core element of the above mentioned producer association is the preservation of the endangered, traditional Swabian–Hall swine pig breed. Accordingly, the animals must come from the district Swabian–Hall, Hohenlohe, Main-Tauber, Ansbach, Ostalb, or Rems-Murr. Furthermore, all animals must be slaughtered at the producer slaughter house in Swabian–Hall [5]. The registration of “Swabian–Hall Quality Pork” as a protected geographical indication (PGI) in Europe in conjunction with a specification regulating production and origin was another milestone in marketing [6]. PGI of agricultural and food products requires that at least one stage of the production process is performed within the defined geographical area [7,8,9,10]. Today, only about 1500 sows are registered for this breed. Without exception, these come from farms that belong to the Farmer Producer Association of Swabian Hall, which uses a control system that strictly monitors the quality of the feed given to the animals. Due to the higher production costs, prices for Swabian–Hall pork are about 20–30% higher than prices for “regular” pork depending on the amount of the average market price for pork (range between 1.10 and 1.50 EUR/kg), on the individual cuts, and on the level of processing [2, 11]. The increased demand and higher market prices make Swabian–Hall Quality (SHQ) pork an interesting commodity for food fraud. Conventionally produced pork meat can easily be mislabeled, because SHQ pork cannot easily be distinguished from conventionally produced pork. Thus, analytical methods are required to detect or prevent this potential fraud.
In the past, there have been some approaches to differentiate between meat from different species or geographical origin. These studies used a combination of 1H NMR spectroscopy (to obtain a “chemical fingerprint” of each sample) and multivariate statistical analysis, such as principal component analysis (PCA) and linear discriminant analysis (LDA) [12, 13]. Here, 1H NMR spectroscopic analysis of the non-polar and polar metabolites of pork meat with multivariate data analysis was applied to distinguish between conventionally produced pork (non-SHQ pork) and SHQ pork produced according to binding producer guidelines for a controlled and species-appropriate production.
Materials and methods
Meat samples
A total of 285 raw meat samples were collected between 2018 and 2021. Most of the samples were from the state of Baden-Württemberg, Germany, and were taken by official food inspectors of the German Federal State of Baden Württemberg and the Farmer Producer Association of Swabian Hall. Other samples were obtained from local supermarkets and butchers. The samples included 180 non-SHQ pork samples and 105 SHQ pork samples. The samples included a variety of cuts as well as mince. A total of 275 meat samples (175 non-SHQ and 100 SHQ) were used for multivariate data analysis and establishment of the model. The remaining 10 samples were used for external validation (5 non-SHQ and 5 SHQ). Table S1 provides an overall summary of the samples used. Information on sex, cut and origin can be found.
Chemicals
All reagents and standard compounds were of analytical or high-performance liquid chromatography grade. Sodium chloride, sodium dihydrogen phosphate (≥ 99.0%), 3-(trimethylsilyl)-propionic acid-d4 sodium salt (TSP, 98.0% atom % D), D2O (99.9% atom % D), methanol-d4 (99.9% atom % D), methanol (≥ 99.9%), and chloroform (≥ 99.8%) were obtained from Merck (Darmstadt, Germany). Deuterated chloroform-d1 (≥ 99.8% atom % D) and the internal reference standard tetramethylsilane (TMS) were from Roth (Karlsruhe, Germany).
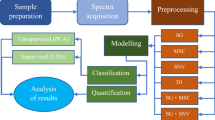
Sample preparation
Meat samples were freed from bones, rind, subcutaneous fat, and innards, mixed, freeze-dried, and ground in a cryomill (SamplePrep6870 Freezer Mill, C3 Process and Analysis Technology GmbH, Haar, Germany). Samples were stored at − 20 °C until being used.
Aqueous extraction Meat powder (500 mg) was extracted with 6 mL of water. After the samples were mixed on a test tube shaker (Multi Reax, Heidolph, Schwabach, Germany) for 10 min, samples were centrifuged at 3000 rpm (relative centrifugal force (RCF), 1690×g) for 15 min. The aqueous layer was passed through a syringe filter (Chromafil Xtra PET − 45/25, Macherey–Nagel, Düren, Germany) into a centrifuge tube. The 3 kDa ultrafiltration filter (Vivaspin®, Sartorius, Göttingen, Germany) was rinsed three times with 2 mL of water each to remove glycerol (centrifugation for 10 min at 3000 rpm, relative centrifugal force (RCF), 1690×g). After glycerol removal, 800 µL of the meat filtrate were transferred to the 3 kDa ultrafiltration filter, followed by centrifugation at 3000 rpm (relative centrifugal force (RCF), 1690×g) for 1.5 h. An aliquot of the filtrate (500 µL) was mixed with 250 µL of 3 M sodium dihydrogen phosphate buffer (pH 6), 75 µL of TSP (dissolved in deuterium oxide, 0.06 M). Finally, a 600 µL-aliquot was transferred to a 5-mm Boro 300–5-8 (Deutero, Bad Kreuznach, Germany) NMR tube.
Fat extraction The meat powder (3 g) was extracted with 20 mL of a mixture of chloroform, methanol, and water (10:5:1, v/v/v). To improve mixing, glass beads were added, and the samples were mixed on a test tube shaker (Multi Reax, Heidolph, Schwabach, Germany) for 15 min. The suspension was passed through a filter (150 mm; Macherey Nagel, Düren, Germany), and 2 mL of 0.9% sodium chloride solution was added to the filtrate in a separation funnel. After three times shaking and a waiting time of 20 min complete phase separation was achieved. The organic phase was vaporised at 45 °C under a stream of nitrogen. An aliquot of the obtained lipid fraction (35 mg) was dissolved in 700 µL of a mixture of chloroform-d1 (containing 0.5% TMS) and methanol-d4 (3:2, v/v); 600 µL of the solution were transferred into a 5-mm Boro 300–5-8 NMR tube.
NMR measurements
All 1H NMR spectra were acquired on a Bruker 400 MHz AVANCE III HD NanoBay spectrometer (Bruker Biospin GmbH, Reinstetten, Germany) equipped with a 5-mm BBI (broadband inverse) probe and a Bruker automatic sample changer Sample Xpress. Temperature equilibration for each sample was 5 min.
Analysis of the aqueous extracts All 1H NMR spectra were recorded using a standard Bruker pulse program noesygppr1d_d7.eba with a relaxation delay (D1) of 4 s and an acquisition time of 8 s. The one-dimensional NMR experiment was performed at 300 K with 128 k time domain data points, 128 scans, 4 dummy scans, a spectral width of 20.5617 ppm, and a receiver gain of 64.
Analysis of the non-polar extracts All 1H NMR spectra were recorded using a standard Bruker pulse program zg30 with a relaxation delay (D1) of 4 s and an acquisition time of 8 s. The one-dimensional NMR experiment was performed at 290 K with 128 k time domain data points, 128 scans, 2 dummy scans, spectral width of 20.0024 ppm, and a receiver gain of 45.2.
Processing The free induction decays obtained of both methods were processed with Bruker Biospin Topspin software (version 3.2). An exponential window function was applied, and line broadening was set to 0.3 Hz, followed by a Fourier Transformation, spectral phasing, and baseline correction. Spectra were referenced to the TMS or TSP signal at 0 ppm.
All spectra were recorded under the same conditions. To ensure the quality of the spectra, the full width at half maximum of the internal reference signals (TSP and TMS) was determined. A limit of 1.2 Hz was set; if this was exceeded, the measurement or sample preparation had to be repeated.
Chemometric methods
Data reduction and pretreatment of the 1H NMR spectra Bucketing was performed for data reduction and to provide the input variables for the following statistical analysis.
Analysis of the non-polar extracts The spectral region 0.50–9.50 ppm was divided into 1000 equal segments, and the region around the signal of residual water (4.84–5.10 ppm) was excluded. Spectra were normalised to the signal of TSP (− 0.5 to 0.5 ppm). A pseudo-scaling effect was achieved by log transformation [14, 15].
Analysis of the non-polar extracts The spectral region 0.50–6.8 ppm was divided into 2000 equal segments, and the regions around the methanol (3.32–3.41 ppm) and residual water (4.46–4.80 ppm) signals were excluded. Spectra were normalised to the signal region 1.50–4.25 ppm. Again, a pseudo-scaling effect was achieved by a log transformation [14, 15]. NMR data were analysed using MATLAB version 2019b (The Math Works, Natick, MA, USA).
Multivariate statistical data analysis The potential to differentiate pork meat using NMR data was validated using a combination of established multivariate statistical tools including PCA with LDA and multivariate analysis of variance within a cross-validation (CV) embedded in a Monte Carlo (MC) resampling approach [16, 17]. As classification rule, a test set object was assigned to the class with minimum distance between test set object and respective class mean, that is, assignment according to the nearest class mean (NCM) [18].
Model building PCA/LDA and MC embedded CV (MCCV) 275 meat samples were used to build and validate the prediction model. 90% of these samples were used to build the model, and 10% of the samples were left out as an internal test set. A PCA was performed to reduce the dimensions followed by LDA to get a maximum of class separation [19, 20]. The quality was assigned using the NCM. The distance between the object of the test set and the class means of the model set was compared and the group membership was assigned. To validate the predictivity of the PCA/LDA, a CV with ten randomly selected disjunct subsequent test sets was performed. To avoid any segmentation bias, CV was repeated 10 times with an MC resampling approach (MC = 10) with a new random segmentation for each CV step (i.e., tenfold randomised tenfold cross-validation). Finally, the rates of correct and false class predictions were calculated for each class to set up a confusion matrix.
Identification of possible marker compounds responsible for discrimination The PCA/LDA score and loading plots were plotted using the MATLAB version 2019b. By interpretation of the loading plots, variables were extracted that mostly affect the discrimination or separation in the score plot.
Mid-level data fusion Fusion of 1H NMR data of both the aqueous and non-polar extracts was investigated with a mid-level data fusion approach using MATLAB version 2019b with Statistical Toolbox. First, the data sets consisting of 275 samples were separately subjected to data pretreatment (bucketing, selection of relevant metabolites performed by solvent exclusion, normalization, and log transformation). PCA was then used to perform data reduction from each data matrix, resulting in the respective scores. In the next step, the PCA scores were fused, resulting in a joint data set. This data set was used to perform the LDA [21,22,23].
Results and discussion
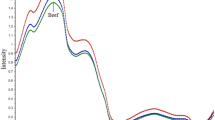
NMR spectroscopy allows for the evaluation of hundreds of chemical compounds in a single experiment [18]. For the investigation of raw muscle meat by NMR, there are already studies that have identified numerous polar and non-polar metabolites. Castejón's study identified 60 metabolites in an aqueous beef extract, 23 of them for the first time in NMR meat studies [12, 13, 28]. Representative one-dimensional 1H NMR spectra of aqueous and non-polar (lipid) extracts of SHQ pork and non-SHQ pork are shown in Fig. 1. Meat fat is composed of two different major lipid classes: triglycerides as neutral lipids and charged phospholipids. Cholesterol and free fatty acids are also present in comparably low concentrations [25, 26]. Polar metabolites of meat include numerous low molecular weight compound classes of high chemical diversity. The main classes are amino acids, organic acids, carbohydrates, purine derivatives, imidazole dipeptides, quaternary ammonium compounds, and amino acid derivatives [28]. Closer inspection of the spectra (Fig. 1) reveals differences in the metabolite composition between SHQ pork and non-SHQ pork. As for example, in the aqueous extracts, the signal of α-alanine at 1.48 ppm shows differences between the two groups. In addition, differences at 2.1–2.2 ppm and 2.3–2.4 ppm can be seen; within this range signals of the amino acids glutamine and glutamic acid can be found. In the low-field range, differences can be seen at 7.23 and 8.50 ppm, which can be attributed to the imidazole dipeptides anserine and carnosine. In the non-polar extract, differences in phospholipid composition are demonstrated by the signal range 3.5–4.5 ppm.
Classification of pork meat by 1H NMR spectroscopy and combined multivariate statistical analysis
Figure 2 shows the results of the embedded MCCV as a confusion matrix of the obtained classification model for the non-polar (A) and polar (B) metabolites of the pork samples. For the LDA of data obtained from the non-polar metabolites, the scores of the first 15 dimensions of the PCA were used, describing 96.5% of the total variance of the data. The confusion matrix, in which classification accuracies are given in percent, demonstrates that the obtained model is suitable for the differentiation of SHQ pork and non-SHQ pork by analysis of the lipophilic metabolites: the accuracy of assignment to the correct class is between 91.4 and 94.0%. The vast majority of non-SHQ pork samples (160 samples of 175 samples) were assigned to the correct class using the embedded MCCV. A total of 13 non-SHQ samples were assigned to the false class in the entire MCCV, that is, in each MC run. Two non-SHQ pork samples were misclassified only twice. Considering the SHQ pork samples, 94 out of 100 samples were assigned to the correct class, whereas six samples were assigned to the wrong class in the entire MCCV.
Results of embedded Monte Carlo cross-validation (MCCV) with respect to the performance of the obtained PCA–LDA based classification model for the prediction of non-SHQ pork (blue) and SHQ pork (turquoise). The analysed data were obtained from 1H NMR spectra of the non-polar metabolites (A) and polar metabolites (B). The left figures in A and B show the confusion matrices of the MCCV. The x-axis and the y-axis show the assigned class and the correct class. The confusion matrices provide information about false and true positive and negative sample classifications, respectively, given in percent. The figures on the right side in A and B show the discrimination space of a single cross-validation step. The training set (indicated as circles) for model building of each class is symbolised by its 95% confidence ellipsoid, and the test set samples are marked as squares. Samples that were incorrectly assigned are marked as red asterisks
In the cloud model, the two groups are shown in a 95% confidence interval. The two groups of samples overlap slightly in the cluster model, which shows the first three linear discriminant functions. Figure 2A (bottom right) also shows that the majority of the 21 samples that were assigned to the incorrect class are located in the overlapping area of the two clusters (red stars).
Figure 2B shows the results of the embedded MCCV and the obtained classification model based on the polar metabolites. The scores of the first 15 dimensions of the PCA, which describe 93.9% of the total variance of the data, were used for LDA. The confusion matrix demonstrates that the model that is based on the polar metabolites is also suitable for the differentiation of SHQ pork and non-SHQ pork: the accuracy of assignment to the correct class is between 90.0 and 94.8%. Using embedded MCCV, 166 of 175 non-SHQ samples were correctly classified. Five non-SHQ pork samples were assigned to the false class in all MC runs. The remaining non-SHQ pork samples were misclassified only once. Accordingly, 90 out of 100 SHQ pork samples were assigned to the correct class, whereas six samples were assigned to the false class in the entire MCCV. Each of the remaining four samples was assigned to the non-SHQ class three times.
Just as shown for the non-polar metabolites, the two groups of samples form clusters that overlap slightly (Fig. 2B, top right). In addition, the scatter of the muscle meat samples differs within the two clusters. The SHQ pork samples scatter more distinctly than the non-SHQ samples. Again, most of the 19 samples (red stars, Fig. 2B, bottom right) that were incorrectly assigned are located in the overlapping area of the two clusters.
Classification-relevant metabolites
Spectral regions of the 1H NMR spectrum that contain buckets with the greatest impact on the clustering of the respective sample set can be extracted from PCA/LDA loading plots (Fig. 3). Using the loadings, it was possible to identify signal regions in the 1H NMR spectra of the non-polar metabolites that were responsible for the separation of the clusters (Fig. 3A) [20, 24]: the buckets in the range 3.212/3.218 ppm are associated with the head group of phosphatidylcholine. These buckets correlate with negative score values of the non-SHQ pork group along LD 1 and, therefore, hold information for this group. In addition, the buckets at 4.006 and 3.596 ppm have high negative loading values and also reflect phospholipids. The buckets at 5.368/5.375 ppm correspond to protons involved in a double bond and thus represent unsaturated fatty acids. Furthermore, a cluster of numerous buckets is found in the region of 2.034 and 2.833 ppm. The former correspond to the signal of the methylene group adjacent to a double bond, whereas the latter represent polyunsaturated fatty acids (PUFA). Thus, it can be concluded that the contents of unsaturated fatty acids, especially PUFA, and phospholipids are higher in non-SHQ pork as compared to SHQ pork. In contrast, the high positive loading values along LD 1 correlate with the SHQ pork samples. Here, buckets were identified in the region of 1.257/1.263/1.269 ppm. These buckets represent the signal region of fatty acid methylene groups. Furthermore, the region at 0.885 ppm exhibits a positive loading value along LD 1. The signals of terminal methyl groups of fatty acids are found in this region.
A Two-dimensional PCA/LDA score plot of the sample group SHQ pork (turquoise) and non-SHQ pork (blue) and associated loading plot with the 1832 buckets used. The buckets with the highest positive or highest negative values along LD 1 are marked and correspond to signal regions in the 1H NMR spectrum of the non-polar metabolites that are more distinct in the respective sample group. B Two-dimensional PCA/LDA score plot of the sample group SHQ pork (turquoise) and non-SHQ pork (blue) and associated loading plot with the 967 buckets used. The buckets with the highest positive or highest negative values along LD 1 are marked and correspond to signal regions in the 1H NMR spectrum of polar metabolites that are more distinct in the respective sample group
These data suggest that SHQ pork and non-SHQ pork vary in their lipid composition. The fat fraction of SHQ pork is richer in saturated fatty acids and the percentage of phospholipids in the total fat is lower; thus, the percentage of triglycerides appears to be higher. In general, the content of phospholipids in muscle meat is lower than the triglyceride content. Moreover, it remains relatively constant independent of the total fat content (lean vs. high-fat). However, the proportion of phospholipids in relation to the total lipid content can change due to the increased amounts of triglycerides [25]. Thus, if the degree of fatness of an animal increases, primarily only the fraction of triglycerides changes. The relative amount of phospholipids can vary from 10 to 50% (based on total lipid content) and depends on factors, such as the species, age of the animal, and feeding [26, 27]. Thus, the lipid fraction of non-SHQ samples is richer in unsaturated fatty acids such as PUFA and the proportion of phospholipids in relation to total fat content is higher. In addition, in the literature, SHQ pork has been described as being particularly rich in fat [1, 3, 4].
For the polar metabolites, buckets which are relevant for class separation were identified in the range of 3.40–4.00 ppm (Fig. 3B). Buckets in the range of 3.410–3.440 ppm showed high positive loading values for the non-SH pork samples. Since signals of a variety of metabolites such as α/β-glucose, proline, and taurine can be found in this range, an unambiguous identification was not possible [28]. For SHQ pork samples, the highest negative loading values for the buckets were obtained at 3.635, 3.374 ppm, and 4.095 ppm. Again, unambiguous metabolite identification was not possible due to numerous signal overlaps.
Mid-level data fusion
Because the clusters slightly overlap in both classification models, a mid-level data fusion was performed using the 275 spectra of each, the non-polar and polar metabolites. Figure 4 describes the LDA based on the merged PCA scores of the two extraction methods. The classification accuracy, which was between 90 and 95% for the individual models, was improved by data fusion. By combining the data, the accuracy of assignment to the correct class is 97.3 and 98.3%, respectively. Furthermore, the two group clusters do not overlap in the cloud model based on a confidence interval of 95%. As a result, less samples were incorrectly assigned. In total, only two samples were assigned to the wrong class in the entire MCCV. The remaining three samples were incorrectly assigned only once or twice.
Results of embedded Monte Carlo cross-validation (MCCV) with respect to the performance of the obtained PCA–LDA based classification model for the prediction of non-SHQ pork (blue) and SHQ pork (turquoise) after mid-level data fusion. Mid-level data fusion combined the scores of both methods generated by the previously performed PCA. A new classification model was generated for the prediction of non-SHQ pork (blue) and SHQ pork (turquoise). The left figure shows the confusion matrix of the MCCV. The x-axis and the y-axis show the assigned class and the correct class, respectively. The confusion matrices provide information about false and true positive and negative sample classifications, respectively, given in percent. The figure on the right shows the discrimination space of a single cross-validation step. The training set (indicated as circles) for model construction of each class is symbolised by its 95% confidence ellipsoid, and the test set samples are indicated as squares
The robustness was tested by applying the generated classification model to independent samples. A test set consisting of five non-SHQ samples and five SHQ samples each was used for external validation. The test set samples were not previously involved in the model construction [22]. 1H NMR spectra of the lipophilic and hydrophilic metabolites, respectively, were acquired for the ten independent test samples. For both groups, the test set samples were assigned to the correct class, but two SHQ pork samples (sample 100, sample 101) were outside the 95% confidence ellipsoid (Fig. 5).
External validation results of the classification model after mid-level data fusion (1H NMR data of polar and non-polar metabolites). The discrimination spaces of a single cross-validation step are shown. The training set for model construction of each class is symbolised by its 95% confidence ellipsoid, and the validation set samples are indicated as red asterisks. The table below shows the corresponding p values. For the p value, if the p value is ≤ 0.05, the samples do not meet the specified significance level. A External validation for the SHQ pork class (turquoise), B external validation for the non-SHQ pork class
Conclusion
It is possible to differentiate between SHQ pork and non-SHQ pork using 1H NMR spectroscopy in combination with multivariate data analysis. However, additional research is required to identify the polar metabolites that are responsible for discrimination. The developed methods demonstrate the power of this approach in the analysis of meat authenticity. Potential applications include the differentiation of conventional and organic meat and meat products. In addition, additional studies to differentiate samples with regard to the geographical origin or origin-protected meat products (protected designation of origin or PGI) appear to be promising.
Abbreviations
- CV:
-
Cross-validation
- 1H NMR:
-
Proton nuclear magnetic resonance
- LD:
-
Linear discriminant function
- LDA:
-
Linear discriminant analysis
- MC:
-
Monte Carlo
- NMC:
-
Nearest class mean
- NMR:
-
Nuclear magnetic resonance
- PCA:
-
Principal component analysis
- PGI:
-
Protected geographical indication
- PUFA:
-
Polyunsaturated fatty acids
- SHQ pork:
-
Swabian–Hall Quality pork
- TSP:
-
3-(Trimethylsilyl)-propionic acid-d4 sodium salt
- TMS:
-
Tetramethylsilane
References
Coolsaet B (2015) Transformative participation in agrobiodiversity governance: making the case for an environmental justice approach. J Agric Environ Ethics 28:1089–1104. https://doi.org/10.1007/s10806-015-9579-2
Larson J (2007) Relevance of geographical indications and designations of origin for the sustainable use of genetic resources. Global Facilitation Unit for Underutilized Species, Rome
Pappenberger EM (2014) Vergleich der Körperzusammensetzung konventioneller und alternativer Masthybriden mittels Dualenergieröntgenabsorptiometrie und Magnetresonanztomographie. Dissertation: Ludwig-Maximilians-Universität München, Tierärztlichen Fakultät, A. Scholz
Petig M (2020) Wachstum, Schlachtkörper-und Fleischqualität der bedrohten Schweinerasse Schwäbisch-Hällisches Schwein unter besonderer Berücksichtigung von Haltungs-, Fütterungs-, und Kombinations-kreuzungsvarianten. Dissertation: Justus-Liebig-Universität Gießen, Fachbereich Agrarwissenschaften, Ökotrophologie und Umweltmanagement, S. König
Bäuerliche Erzeugergemeinschaft Schwäbisch Hall (Fassung 2014) Verbindliche Erzeugerrichtlinien für Schwäbisch-Hällisches Qualitätsschweinefleisch g.g.A. aus kontrollierter und artgerechter Erzeugung. Urheberrechtliches Eigentum des Konsortiums Bäuerliche Erzeugergemeinschaft Schwäbisch Hall. https://www.besh.de//images/downloads/Richtlinien_SH_ggA.pdf. Accessed 7 Oct 2021
European Commission, Regulation (EC) No 644/98 of 20 March 1998 supplementing the Annex to Regulation (EC) No 1107/96 on the registration of geographical indications and designations of origin under the procedure laid down in Article 17 of Council Regulation (EEC) No 2081/92. https://eur-lex.europa.eu/legal-content/EN/ALL/?uri=CELEX%3A31998R0644
Albayrak M, Gunes E (2010) Implementations of geographical indications at brand management of traditional foods in the European Union. Afr J Bus Manag 4:1059–1068. http://www.academicjournals.org/AJBM
Council Regulation (EC) No. 510/2006 of 20 March 2006 on the protection of geographical indications and designations of origin for agricultural products and foodstuffs. https://eur-lex.europa.eu/legal-content/EN/TXT/?uri=celex:32006R0510
Council Regulation (EC) No. 2081/92 of 14 July 1992 on the protection of geographical indications and designations of origin for agricultural products and foodstuffs. https://eur-lex.europa.eu/eli/reg/1992/2081/oj
European Commission, Regulation (EC) No. 1151/2012 of the European Parliament and of the Council of21 November 2012 on quality schemes for agricultural products and foodstuffs. https://eurlex.europa.eu/legalcontent/en/TXT/?uri=CELEX%3A32012R1151
Leipprand A, Görlach B, Schlegel S (2006) Case study “Schwäbisch-Hällisches Qualitätsschweinefleisch” (Schwäbisch-Hall quality pork). Ecologic-Institute for International and European Environmental Policy
Jakes W, Gerdova A, Defernez M, Watson AD, McCallum C, Limer E, Colquhoun IJ, Williamson DC, Kemsley EK (2015) Authentication of beef versus horse meat using 60 MHz 1H NMR spectroscopy. Food Chem 175:1–9. https://doi.org/10.1016/j.foodchem.2014.11.110
Jung Y, Lee J, Kwon J, Lee KS, Ryu DH, Hwang GS (2010) Discrimination of the geographical origin of beef by 1H NMR-based metabolomics. J Agric Food Chem 58:10458–10466. https://doi.org/10.1021/jf102194t
Emwas AH, Saccenti E, Gao X, McKay RT, dos Santos VA, Roy R, Wishart DS (2018) Recommended strategies for spectral processing and post-processing of 1D 1H-NMR data of biofluids with a particular focus on urine. Metabolomics 14:1–23. https://doi.org/10.1007/s11306-018-1321-4
Euceda LR, Giskeødegård GF, Bathen TF (2015) Preprocessing of NMR metabolomics data. Scand J Clin Lab Invest 75:193–203. https://doi.org/10.3109/00365513.2014.1003593
Longobardi F, Ventrella A, Napoli C, Humpfer E, Schuetz B, Schaefer H, Kontominas MG, Sacco A (2012) Classification of olive oils according to geographical origin by using 1H NMR fingerprinting combined with multivariate analysis. Food Chem 130:177–183. https://doi.org/10.1016/j.foodchem.2011.06.045
Godelmann R, Fang F, Humpfer E, Schuetz B, Bansbach M, Schaefer H, Spraul M (2013) Targeted and nontargeted wine analysis by 1H NMR spectroscopy combined with multivariate statistical analysis. Differentiation of important parameters: Grape variety; geographical origin; year of vintage. J Agric Food Chem 61:5610–5619. https://doi.org/10.1021/jf400800d
Ackermann SM, Lachenmeier DW, Kuballa T, Schuetz B, Spraul M, Bunzel M (2019) NMR-based differentiation of conventionally from organically produced chicken eggs in Germany. Magn Reson Chem 5:579–588. https://doi.org/10.1002/mrc.4838
Yi L, Dong N, Yun Y, Deng B, Ren D, Liu S, Liang Y (2016) Chemometric methods in data processing of mass spectrometry-based metabolomics: a review. Anal Chim Acta 914:17–34. https://doi.org/10.1016/j.aca.2016.02.001
Trygg J, Holmes E, Lundstedt T (2007) Chemometrics in metabonomics. J Proteome Res 6:469–479. https://doi.org/10.1021/pr060594q
Borràs E, Ferré J, Boqué R, Mestres M, Aceña L, Busto O (2015) Data fusion methodologies for food and beverage authentication and quality assessment—a review. Anal Chim Acta 891:1–14. https://doi.org/10.1016/j.aca.2015.04.042
Callao MP, Ruisánchez I (2018) An overview of multivariate qualitative methods for food fraud detection. Food Control 86:283–293. https://doi.org/10.1016/j.foodcont.2017.11.034
Smolinska A, Engel J, Szymanska E, Buydens L, Blanchet L (2019) Chapter 3—general framing of low-, mid-and high-level data fusion with examples in the life sciences. In: Cocchi M (ed) Data fusion methodology and applications, vol 31, 1st edn. Elsevier, Amsterdam. https://doi.org/10.1016/B978-0-444-63984-4.00003-X
Wold S, Esbensen K, Geladi P (1987) Principal component analysis. Chemometr Intell Lab Syst 2:37–52. https://doi.org/10.1016/0169-7439(87)80084-9
Wood J, Enser M, Fisher AV, Nute GR, Sheard PR, Richardson RI, Hughes SI, Whittington FM (2008) Fat deposition, fatty acid composition and meat quality: a review. Meat Sci 78:343–358. https://doi.org/10.1016/j.meatsci.2007.07.019
De Smet S, Raes K, Demeyer D (2004) Meat fatty acid composition as affected by genetic factors. Anim Res 53:81–88. https://doi.org/10.1051/animres:2004003
Enser M, Hallett KG, Hewett B, Fursey GAJ, Wood JD, Harrington G (1998) Fatty acid content and composition of UK beef and lamb muscle in relation to production system and implications for human nutrition. Meat Sci 49:329–341. https://doi.org/10.1016/s0309-1740(97)00144-7
Castejón D, García-Segura JM, Escudero R, Herrera A, Cambero MI (2015) Metabolomics of meat exudate: its potential to evaluate beef meat conservation and aging. Anal Chim Acta 1:1–11. https://doi.org/10.1016/j.aca.2015.08.032
Acknowledgements
The authors thank Cedric Kunz for excellent technical assistance. In addition, we thank the Farmer Producer Association of Swabian Hall for providing authentic sample material for this study.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there are no conflicts of interest.
Compliance with ethics requirements
This article does not contain any studies with human or living animal subjects.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Decker, C., Krapf, R., Kuballa, T. et al. 1H NMR spectroscopy combined with multivariate data analysis for authentication of “Swabian–Hall Quality Pork” with protected geographical indication. Eur Food Res Technol 249, 2559–2567 (2023). https://doi.org/10.1007/s00217-023-04312-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00217-023-04312-y