Abstract
This paper presents the effect of polyphenols on microorganisms inhabiting the human gastrointestinal tract (mainly bacteria belonging to the Lactobacillus genus) and pathogenic microorganisms classified as the most common food contaminants. Plant secondary metabolites have the ability to modulate the growth of many microorganisms. Due to the metabolic changes induced by their presence in the environment, many pathogenic microorganisms are unable to grow, which in turn cause a significant reduction in their pathogenic potential. These processes include primarily the induction of ruptures in the cell membrane and disturbance of cell respiration. Often, the lack of integrity of cell membranes also leads to the disturbance of intracellular homeostasis and leakage of cellular components, such as proteins, ATP molecules or intracellular ions. Autoxidizing polyphenols also act as pro-oxidative substances. Hydrogen peroxide formed in the process of oxidation of polyphenolic compounds acts as a bactericidal substance (by induction of DNA breaks). With regard to intestinal microbiota, polyphenols are considered prebiotic substances that increase the number of commensal bacteria. They can positively influence the growth of Lactobacillus bacteria, which have the ability to metabolize undigested antioxidants in the digestive tract of humans and animals. Depending on the pH of the environment and the presence of ions, plant polyphenols in the human digestive tract can act as substances with antioxidant potential or become pro-oxidants. Thus, combining functional food with polyphenols and Lactobacillus bacteria not only protects food products against the development of undesirable and pathogenic microbiota, but also has a positive effect on human health. The paper also describes the possibility of changes in the genome of Lactobacillus bacteria (under the influence of polyphenols) and the influence of Lactobacillus spp. bacteria on the antimicrobial properties of polyphenols. The enzymatic abilities of bacteria of the genus Lactobacillus, which influence the transformation of polyphenolic compounds, were also described.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Polyphenols, as secondary metabolites of plants, comprise a large group of organic compounds. Their numerous properties, including their antioxidative and antimicrobial (against fungi and bacteria) activity, have been examined by scientists in various fields of science, i.e. medicine, biochemistry, epidemiology and pharmacy [1]. These compounds are particularly abundant in leguminous plants, nuts, cereals, vegetables and fruit. Animal bodies are not capable of synthesizing polyphenols. Instead, they obtain them entirely from the consumption of foods of plant origin [2]. The basic division of polyphenols is based on their molecular structure, i.e. the number of aromatic rings included in their structure. Moreover, this division is based on the types of bonds present between consecutive rings. From this division, four main polyphenol classes, i.e. phenolic acids, stilbenes, lignans and flavonoids (catechins, flavones, isoflavones, flavanones, flavanols, anthocyanins) have been derived [3].
The structure of polyphenols makes it possible to distinguish among them low-molecular (phenolic acids) and high-molecular weight (tannic acids) compounds. Owing to their varied structure, polyphenols demonstrate various physical, biological and chemical properties [4]. The structure of polyphenols, based on active phenolic hydroxyl groups (up to a few groups in a single molecule), gives them antioxidative properties, and has considerable influence on their ability to neutralize free radicals [5]. A polyphenol’s activity depends largely on the number of hydroxyl groups included in an individual molecule [6].
Antagonistic activity of polyphenols
Polyphenols possess a broad range of antagonistic activity towards pathogenic microbiota. For example, polyphenols isolated from tea are able to limit the multiplication of such pathogens as Staphylococcus aureus, Salmonella Enteritidis, Serratia marcescens and Cronobacter sakazakii [7,8,9]. However, during tests performed by Tabasco et al. [10] it was determined that a mixture of catechin, epicatechin and flavan-3-ol, in concentrations of 0.25–1.0 mg/ml, is capable of inhibiting the development of such pathogenic species as Staphylococcus aureus, Streptococcus pyogenes, Staphylococcus epidermidis and Enterococcus faecalis. Piekut [11] demonstrated that lovage extracts rich in anisic, cinnamic, ferulic, sentisic, syringic and p-coumaric acids, after 24-h incubation, inhibited the multiplication of Escherichia coli by 93% and Staphylococcus aureus by 87%. On the other hand, a 48-h culture in the presence of the extract resulted in the inhibition of the development of Pseudomonas aeruginosa and Bacillus subtilis bacteria, as well as Candida albicans yeast by over 90%. Moreover, thyme extract rich in anisic, gallic, sentisic, caffeic, p-coumaric, syringic and vanillic acids inhibited the multiplication of only two pathogenic bacterial strains, Staphylococcus aureus and Escherichia coli. Following a 48-h incubation period, the multiplication of the above-mentioned bacteria was reduced by over 80%.
The basic feature of polyphenols (and their metabolites) responsible for their impact on microorganisms is their chemical structure, including the substituents on the phenolic ring. These features result in the fact that, depending on their concentration, polyphenols can act both as activators and inhibitors of bacterial multiplication and development [12].
Bacterial cell wall integrity disruption by polyphenols
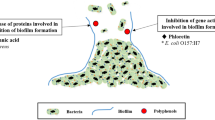
The results of current studies show a clear tendency for Gram-positive bacteria to be much more susceptible to the presence of polyphenols than Gram-negative bacteria [13]. It is believed that Gram-negative bacteria are able to be more resistant to the bactericidal activity of polyphenols due to the structure of their cell walls and the arrangement of their external membranes. This resistance results from the fact that the Gram-negative bacteria structure includes periplasmic space which is not present in Gram-positive bacteria. Moreover, the periplasmic space is rich in enzymes capable of degrading compounds demonstrating antimicrobial potential, permeating to the external environment [14]. The external membrane of Gram-negative bacteria cells is rich in lipopolysaccharides, creating a barrier that is practically impermeable for lipophilic molecules. Active components are able to bond with the cell surface in order to be subsequently transported inside. The effectiveness of polyphenols consists in reaching the target location, i.e. phospholipid cell membranes, or in altering the synthesis of intracellular compounds, i.e. enzymes, ATP or intracellular proteins [15]. In most cases, polyphenols demonstrate a hydrophobic character, because of which they readily interact with the lipid layer located in the bacterial cytoplasm membrane. Owing to their integration with the double lipid layer of the cytoplasm membrane, polyphenols greatly disturb its stability [16]. This disruption occurs because phenolic acids are capable of causing irreversible changes in cell membrane structures and disturbing their hydrophobic properties. They are also able to induce spot ruptures compromising the impermeability of structures. Pores created in this manner become locations for the permeation of cellular material into the environment [17]. The deformation of a cell membrane’s physical structure caused by the presence of polyphenols may result in the membrane swelling, which in turn results in structural instability and increased passive permeability [18]. As a result of cell membrane loosening, apart from ATP, a bacterium cell is deprived of ions, nucleic acids and amino acids which are necessary for its existence [19] (Fig. 1).
Impact of polyphenols on bacterial virulence
As regards certain pathogens (even those counted as Gram-negative bacteria) that colonize the human body (e.g. Escherichia coli that cause urinary system infections), some polyphenols, such as flavone-3-ol (condensed tannic acid), are able to disturb the expression of genes that are necessary to synthesize p-fimbriae proteins. As a result of the molecule conformation change, Escherichia coli bacteria are deprived of their ability to adhere to the epithelium surface [20]. When uropathogenic Escherichia coli bacteria do not have the ability to bond adhesively with receptors located on the host's membranes, no infection of the mucilaginous epithelium surface and subsequent development of bacterial infection can occur. Plant polyphenols with antimicrobial properties often become a factor that limits the occurrence of bacterial biofilm. Proanthocyanidins (PAC) from cranberry extract inhibit the generation of fructosyl transferase and glucose transferase. Inhibition of the activity of the above-mentioned enzymes ensures that the Streptococcus genus is unable to generate biofilm dependent on sucrose, which results in it being unable to settle on biotic surfaces (e.g. dental plaque). Additionally, PACs derived from cranberry are also able to inhibit biofilms whose generation does not depend on sucrose concentration. Changes in bacterial cell co-aggregation, caused by an increase in the hydrophobic nature of cell walls and modifications on their surface, prevent accumulation of bacterial cells into larger aggregates [21]. It is believed that polyphenols may have properties which alter the adhesion of pathogenic cells (including Escherichia coli) to human epithelium walls. Phytocompounds may influence the host’s cells analogously to competitive receptors and make it impossible to create receptor–ligand bonds (common in the case of urinary system infections) [22]. Alteration of pathogenic cell adhesion in the presence of polyphenols is also visible in Helicobacter pylori. Derivatives of flavan-3-ol demonstrate anti-adhesive properties in relation to the bacteria, which cause inflammation of the stomach mucous membranes [23].
Alterations of bacterial metabolism caused by polyphenols
One of the bactericidal properties of polyphenols is the ability to make alterations on the level of extra- and intracellular proteins. Polyphenols isolated from sugar beet are able to disturb the synthesis and expression of genes responsible for the synthesis of proteins in Staphylococcus aureus, Escherichia coli, Listeria monocytogenes and Salmonella Typhimurium. During an experiment performed by Chen et al. [24], SDS-PAGE analysis demonstrated changes in the level of intracellular proteins for the above-mentioned bacterium species. Due to the increase in the permeability of the cell membranes of these bacterial species, more than 80% of all intracellular proteins were detected in the cell-free supernatant. This proves that polyphenols significantly affect the integrity of bacteria cell membranes. By causing their destabilization, they contribute to the death of bacterial cells. In addition, the above-mentioned experiment also proved that polyphenols penetrating the cell membranes of Escherichia coli affect the expression of bacterial genes coding intracellular proteins. The photo of the gel after SDS-PAGE shows the loss of the ability to synthesize many intracellular proteins in Escherichia coli in culture with the addition of polyphenols from sugar beet molasses (at a concentration of 10 mg/ml). These studies confirm the results obtained by Zhao et al. [25], who presented the effect of sugar cane polyphenols on changes in intracellular protein content in food contaminating bacteria (Escherichia coli, Listeria monocytogenes, Staphylococcus aureus and Salmonella Typhimurium). A similar effect was also observed by Wang et al. [26] after adding Listeria monocytogenes, Escherichia coli and Salmonella Enteritidis lactic acid bacteria to the culture. Lactic acid penetrating inside the bacterial cells affects the physiology and morphology of bacterial cells, causing their degradation. The result of this experiment was also confirmed by tests conducted by Zeng et al. [27]. Moreover, polyphenols are able to damage the structure of intracellular proteins. It is supposed that the fairly rapid process of the decrease in intracellular protein concentration is caused by changes in the metabolism of energy. Changes in protein concentration also induce rebuilding of the cytoplasm membrane and membrane protein structures [28]. The antimicrobial potential of polyphenols mostly results from changes in the intracellular concentration of ATP. Changes in ATP concentration are connected with the depolarisztion of cell membranes, whose disturbed structure causes leakage of cytoplasm (increase in cell membrane permeability) and loss of the ATP molecules necessary for existence [29].
Antioxidative activity of polyphenols
Antioxidants are substances which, at a low concentration with respect to an oxidizing substrate, significantly delay or prevent the oxidation of other compounds by this substrate [30]. In the context of biological processes, the mechanism of the action of antioxidants is more complex and depends on more factors. Antioxidant properties may be based on inhibition of enzymes that produce oxidants or chelation of metal ions mediating oxidation reactions. There may also be a situation in which the antioxidant is oxidized instead of the substrate [31].
During aerobic respiration, bacteria produce oxygen (O2) which is necessary for the production of cellular energy. In the case of incomplete reduction of O2 in the process of cellular respiration, it is possible to generate reactive oxygen species, with particular emphasis on hydrogen peroxide (H2O2) and hydroxyl radicals (OH-). The system of bacteria protection against reactive oxygen species (ROS) is related primarily to their ability to synthesize specific enzymes such as catalase and superoxide dismutase. The addition of polyphenols to bacterial culture usually induces cell lysis, which results in an increase in the production of ROS and H2O2, which results in cell death [32].
Research on the mechanisms of the antimicrobial activity of polyphenols led to the conclusion that this phenomenon may be associated with the high antioxidant potential of these compounds [33]. Antioxidants such as polyphenols are capable of inducing physicochemical and structural changes in microorganisms, thus significantly delaying their growth [34].
To date, two basic mechanisms of the antimicrobial action of polyphenols related to their antioxidant activity have been proven. First of all, by direct production of H2O2 and reduction of Fe3+ ions to Fe2+ (Fenton reaction), they participate in the formation of strong, reactive oxygen species (such as hydroxyl radicals). Nevertheless, a key factor in determining whether antioxidants have antimicrobial properties is the pH value [35].
Hydrogen peroxide is produced by the oxidation of phenolic compounds such as gallic acid, (−)-epigallocatechin, (−)-epigallocatechin gallate, (+)-catechin, quercetin, hydroxytyrosol, delphinidin and rosmarinic acid. H2O2 (in a concentration above 100 µM) is also produced in media with the addition of catechins from green and black tea, coffee or wine. Polyphenols are also able to induce synthesis of hydrogen peroxide in beverages. In studies conducted by Grzesik et al. [31], the most active antioxidants include polyphenols such as propyl gallate (PG), pyrogallol, (−)-epigallocatechin gallate (EGCG) and quercetin (Q). Hydrolysis of hydrogen peroxide, during which oxygen is produced, significantly accelerates the processes of self-oxidation of polyphenols (with simple structures, e.g. coumarin). The antimicrobial activity of polyphenols is possible owing to the loss of antioxidant properties and transformation into a pro-oxidant (a substance that generates free radicals). According to the research carried out by Dai, Mumper [36], it is precisely under conditions favouring self-oxidation (high pH with a simultaneous high concentration of transition metal ions) that polyphenols are transformed into pro-oxidants. Moreover, hydrogen peroxide is a factor necessary to accelerate the process of self-oxidation of polyphenols (through hydrolysis). It is also a source of hydroxyl radicals. The concentration of polyphenols is the primary factor responsible for their antimicrobial properties resulting from the synthesis of hydrogen peroxide. Polyphenols alone cannot induce direct breaks in bacterial DNA. It is H2O2 that disrupts the integrity of the DNA strands [37]. It is believed that hydrogen peroxide, formed in the processes of self-oxidation of polyphenolic compounds (contained in plant extracts and honey), induces DNA strand breaks by accelerating oxidation processes (especially guanine) [38]. The antimicrobial properties of polyphenols may also result from their ability to interact chemically with H2O2. As a result of these reactions, products responsible for the degradation of bacterial DNA are formed [39]. Pro-oxidative polyphenols may cause changes in the expression of bacterial proteins. As a result of interaction with polyphenols in bacteria more susceptible to oxidative stress, DNA damage (as a result of the breakdown of DNA fragments) and, consequently, abnormal transcription processes occur. On the other hand, incorrect transcription can lead to disturbances in the production of superoxide dismutase and catalase (as in the case of damage caused by H2O2). An increased concentration of H2O2 can lead to transcriptional disturbances, as a result of which the genes encoding proteins synthesized during oxidative stress or the genes responsible for enzymes that properly metabolize antioxidants are damaged. A weakened bacterial cell is unable to develop properly, which often leads to its death [40]. On the other hand, while polyphenols continue to function as antioxidants, they may interact with proteins found in the bacterial cell walls. This often leads to the initiation of ionic and hydrogen bonds. Typically, these combinations induce changes in protein activity and a decrease in bacterial resistance. It should be remembered that the antioxidant activity of polyphenols also changes [40]. The mere presence of polyphenols in the growth environment of bacteria (due to their antioxidant potential) causes oxidative stress [35]. Hydrogen peroxide generated by the auto-oxidation of polyphenols may also have an effect not only on microorganisms, but also on cells. Grzesik et al. [31], in studies on DU-145 (human prostate carcinoma) cells, investigated the cytotoxicity of PG, EGCG and Q. The results of their research show that the cytotoxicity of polyphenols is much lower in tests with the addition of catalase (an enzyme that decomposes hydrogen peroxide). Thus, it is likely that the negative impact on cells should not be attributed to polyphenols, but to products formed in the process of their self-oxidation.
It is worth emphasizing, however, that the antioxidant activity (and thus the antimicrobial activity) of polyphenols may depend on many factors. The ability to absorb free radicals depends primarily on the pH of the environment, metal reduction potential, solubility, or activity of chelating polyphenols. The antioxidant properties of polyphenols are observed in systems where plant metabolites use redox active metals (iron, copper) [41, 42].
In studies conducted by Grzesik et al. [31], during the incubation of 54 different substances with antioxidant potential (incubation: 3 hours, 37 °C), as many as 27 antioxidants (mainly natural food ingredients) produced hydrogen peroxide. The kinetics of H2O2 synthesis was linear. The highest concentration of hydrogen peroxide was recorded in the initial phase of incubation (after about 1.5 h). Among the tested antioxidant substances, the highest concentration of H2O2 in the culture media was observed in the case of propyl gallate (PG), (−)-epigallocatechin gallate (EGCG) and quercetin (Q). In these studies, it was also observed that another antioxidant (not derived from plants) may have an influence on the course of antioxidant processes and the production of H2O2. Ascorbate in the environment reduced the production of hydrogen peroxide by 60% for Q and inhibited the production of H2O2 for PG and EGCG. The production of reactive oxygen species for these compounds also depended on light. However, it is not a universal mechanism. In the case of exogenous compounds, incubation in the dark does not weaken the processes of auto-oxidation and the formation of hydrogen peroxide. In the context of microbial culture, it is worth mentioning that the formation of hydrogen peroxide may depend on the composition of the medium. The amount of H2O2 formed as a result of self-oxidation of polyphenols varies significantly in Dulbecco's modified Eagle's medium (DMEM), DMEM supplemented with l-glutamine, or in phosphate-buffered saline (PBS). As a result of PG, EGCG and Q self-oxidation, 110–127 µmol H2O2 was formed in PBS medium, while in DMEM with additional l-glutamine between 41.8 and 157.9 µmol. There are also substrates such as yeast extract peptone dextrose (YPG) in which polyphenols such as EGCG and Q are not oxidized, and thus no hydrogen peroxide is generated. The resulting reactive oxygen species are sometimes the result of the integration of polyphenols with the organic components of the substrate, which initiate the processes of self-oxidation of polyphenols. It is worth remembering that the culture media may be contaminated with transition metal ions. Thus, the amount of hydrogen peroxide formed may also be influenced by the water used to prepare the microbial medium. Tea solutions were tested by Grzesik et al. [31], made using water directly from the tap and based on deionized water. It turned out that in tea based on tap water the concentration of the formed H2O2 was kept at the level of 85 µM, while in the case of deionized water it was only 25 µM. It should be emphasized that the level of contamination with transition metal ions in the culture medium and the water used in the reaction will always be a factor influencing the rate of self-oxidation of polyphenols. It will also indirectly influence the ability to form ROS, and thus it will be a factor influencing the speed of the bactericidal activity of polyphenols.
Unfortunately, in the literature there is no clear correlation between the synthesis of hydrogen peroxide and the chemical structure of polyphenolic compounds. It is presumed that compounds with a simple structure undergo faster autooxidation. It is environmental factors that have a much greater impact on the amount of H2O2 synthesized in the oxidation of polyphenols. Transition metal ions in particular are responsible for catalysing the self-oxidation of compounds with antioxidant potential. From an industrial point of view, it is the contamination of finished products with various transition ions that can have a significant impact on their antioxidant properties. Moreover, polyphenols undergo self-oxidation processes faster, the higher the pH of the environment. For example, ascorbic acid in a solution at a concentration of 10 mM (used as a control for substances with antioxidant potential) generates small amounts of H2O2. By neutralizing the solution to pH 7.0, the amount of H2O2 formed is much higher. Thus, the self-oxidation of polyphenols depends on the pH of the environment. It is assumed that in the human intestine this process may occur much faster than in other sections of the gastrointestinal tract [31, 43]. Therefore, polyphenols may show pro-oxidative activity and inhibit the growth of bacteria in slightly alkaline environments (pH 7.0-8.0) [37].
In their research, Taleb et al. [40], also observed that the osmolarity of date syrup (related to the high sugar content in the syrup) does not have a significant effect on the auto-oxidative activity of polyphenols.
The low concentrations of polyphenols found in plants usually behave as antioxidants, thus, when combined in a nutritional matrix with Lactobacillus spp. bacteria, they can fulfil a protective function [40].
Honeydew honey is an example of a product containing a large amount of phenolic acids and flavonoids, having antioxidant and pro-oxidative properties. The antibacterial activity of honeys (mainly honeydew, less often nectar) is attributed to high concentrations of polyphenols, which may be involved in the production of increased amounts of H2O2 [44, 45].
According to research by Bucekova et al. [37], honeydew honey has the ability to inhibit the multiplication of both Gram-positive (Staphylococcus aureus) and Gram-negative (Escherichia coli) bacteria. However, these properties are more noticeable in Gram-positive bacteria. Thus, in 40% (w/v) solutions of the tested honeydew honeys, the content of hydrogen peroxide was determined after 24-h incubation at 37 °C. The samples contained between 0.3 and 3.4 mM H2O2, and the average value of the H2O2 concentration in the tested samples of honeydew honey was 1.8 mM. Out of 23 tested samples, only two were characterized by poor H2O2 production (less than 1 mM). The total polyphenol content was also determined using the Folin–Ciocalteu method in 20% (w/v) honey solutions. Results are expressed as GAE equivalent (mg/ml) using gallic acid (GAE) as standard. In the tested honey samples, no direct participation of polyphenolic compounds in the antibacterial potential of honey was demonstrated (despite the high content of polyphenols: 30–60 mg GAE/100g honey). However, it has been recognized that phenolic compounds may act synergistically through pro-oxidative effects (producing increased levels of H2O2). Thus, Bucekova et al. [37], to confirm the participation of H2O2 in the antibacterial effect of honeydew honey, subjected 50% (w/v) honey solutions to a 2-h catalase treatment. Samples treated with catalase had lower antimicrobial activity with mean MIC values of 30%, which is evidence that supports the above thesis. According to research by Poli et al. [46], hydrogen peroxide formed as a result of auto-oxidation of polyphenols contained in honey acts as a killer against Pseudomonas aeruginosa bacteria. In order to confirm their thesis, the authors of the above-mentioned study also conducted additional studies. An experiment was carried out in which the hydrogen peroxide in the culture medium was neutralized by adding catalase (1000 U/ml; 2 h). Thus, the addition of the enzyme eliminated the bactericidal effect caused by the polyphenols. In the research conducted by Poli et al. [46], five tested honeys, characterized by high antioxidant capacity: 2.76–8.6 ORAC (µmol/l TE/g of honey) and high content of H2O2 (23-54 µmol/l), have antibacterial properties (plasmid DNA degradation) against the bacteria Pseudomonas aeruginosa. According to research by Taleb et al. [40], hydrogen peroxide was also produced in media with the addition of polyphenol-rich date syrup (605 mg polyphenols/100 g of product), such as tannins, flavonoids, flavonols, anthocyanins, carotenoids. Thus, it has been proven in the research that with the increase in the concentration of date syrup (and thus polyphenols) in the medium, the concentration of H2O2 also increases. The hydrogen peroxide formed in the medium inhibited the multiplication of Escherichia coli and Staphylococcus aureus.
The self-oxidation of polyphenols with the simultaneous synthesis of hydrogen peroxide is therefore considered to be a bactericidal process with a generally beneficial effect on human health [47, 48].
Free radicals resulting from the auto-oxidation of polyphenols can be detected using the spin resonance technique. In studies conducted by Grzesik et al. [31], it was observed that as a result of PG and EGCG self-oxidation, hemiquinone radicals are formed. Thus, the use of the spin resonance technique made it possible to observe that ascorbic acid (one of the main components of lemon juice) significantly reduces the production of H2O2 in tea (acidification of the environment—inhibition of self-oxidation). The process of H2O2 production as a result of oxidation of antioxidant compounds has also been observed in in vivo processes. In a study by Lambert et al. [49], the micromolar synthesis of hydrogen peroxide occurs when the green tea solution is kept in the mouth. Hiramoto et al. [50] found clearly higher concentrations of H2O2 in the urine of coffee drinkers (urinary oxidation of coffee hydroxychromate). It is worth emphasizing, however, that the mechanism of H2O2 production with the use of polyphenols in the culture medium is still a poorly understood process. Often, in vitro studies do not match the in vivo processes [31].
Impact of lactic acid bacteria on polyphenol activity
The research conducted by Smith et al. [51] demonstrated the presence of several different responses of lactic acid bacteria cells to the presence of polyphenols in the environment. These bacteria are able to inhibit activity of proanthocyanidins by modification or degradation of these molecules. Sometimes, in response to polyphenolic substances, bacteria are able to dissociate the polyphenol–substrate complex. Bacteria inhabiting the human digestive system, including Lactobacillus genus bacteria, are able to metabolize plant-derived polyphenols provided along with food. Through catabolic reactions, they modify polyphenol structures [52]. The metabolism of polyphenolic compounds affected by lactic acid bacteria significantly decreases the toxicity level of primary compounds. For example, the metabolized products of such acids as caffeic, p-muramic and protocatechuic acids demonstrate lower antimicrobial potential than the acids in their primary form. However, such factors as the method of polyphenolic compound degradation, the ability ferment them and the possibility of surviving in their presence are individual features of each lactic acid bacteria (LAB) species [53].
Polyphenols present in food have various forms, such as esters, polymers and glycosides. Thus, before they are absorbed in the intestines, these compounds must be hydrolysed so that they can play an active role in the body. This is possible due to the intestinal microbiota [3]. Some probiotic bacteria, such as Lactobacillus plantarum IFPL935, demonstrate high enzymatic activity directed at metabolizing polyphenols. They are capable of synthesizing galloyl-esterase, decarboxylase and benzyl alcohol dehydrogenase. Owing to their demonstrated enzymatic activity, Lactobacillus plantarum IHPL935 bacteria are able to conduct hydrolysis of polyphenols isolated from grape pips. As a result of their reactions, pyrogallol, catechol and gallic acids are generated [10].
Structural modifications of polyphenols caused by the presence of lactic acid bacteria are individual features of each higher organism (human or animal). Thus, the accessibility of certain compounds and their effects are changeable [54]. Lactic acid bacteria can also have a considerable influence on the polyphenolic profile, following the fermentation of juices. According to Kwaw et al. [55], mulberry juice fermented using LAB is characterized by a higher polyphenol content compared to a control sample (raw, unfermented juice). Over 1000 μg/ml of polyphenol compounds can be isolated from raw mulberry juice. Among the secondary metabolites of mulberry juice, syringic acids (31.43% of flavonoids), cyanidin-3-O-rutinoside (51.11% of anthocyanins) and quercetin (39.62% of flavonols) are the most dominant. As a result of juice fermentation using LAB, it is possible to obtain much higher concentrations of polyphenols in a sample. Lactobacillus paracasei, Lactobacillus plantarum and Lactobacillus acidophilus are able to increase the total content of polyphenols by up to 1516, 1661 and 1644 mg/ml, respectively. Similar results were obtained by Pérez-Gregorio et al. [56]. This effect can be related to the ability of LAB to synthesize hydrolytic enzymes, thanks to which it is possible to conduct hydrolyses of complex phytochemical compounds to much simpler structures. However, it must be stressed that the adaptation of lactic acid bacteria to environments containing polyphenols is an individual feature. Within the species, there are also significant differences in the level of hydrolytic enzyme synthesis. Higher concentrations of polyphenols in juice fermented by LAB may also result from polymer degradation, as a result of which complex compounds are transformed into a free form. Polymer degradation is conditioned by phenol oxidases, the synthesis of which depends on LAB [57]. Moreover, the production of lactic acid by LAB bacteria, causing a decrease in the environmental pH value as a result of its increased level, may act on polyphenols as a stabilizing agent. Additionally, polyphenol-rich fruit juice fermentation results in considerable enhancement of its anti-oxidative properties [55]. Undoubtedly, lactic acid bacteria are able to ferment polyphenolic compounds, which results in a direct decrease in the amount of these compounds in plant products. Sunflower flour is a food product characterized by a high content of phenolic acid compounds (up to 4% of the total weight). However, chlorogenic acid (5-O-caffeoylquinic acid), i.e. the ester of caffeic and quinic acids (23–33 g of acid/kg of dry sunflower mass), constitutes 70% of the content of all the polyphenolic compounds that can be isolated from sunflower seeds [58, 59]. Among Lactobacillus genus bacteria, species capable of phenolic acid metabolism, including chlorogenic, caffeic, p-carbamic and protocatechuic acids, were identified [60,61,62]. In certain cases, LAB also exert an impact on the anti-oxidation potential of polyphenols in their environment. For example, the research conducted by Rúa et al. [63] demonstrated that bacteria such as Lactobacillus rhamnosus, Lactobacillus delbrueckii subsp. bulgaricus, Lactobacillus paracasei subsp. paracasei and Lactococcus lactis subsp. cremoris, cause synergistic effects as regards the antioxidative activity of polyphenols contained in melon juice.
Metabolism of polyphenols in the human digestive system
Understanding the processes of polyphenol metabolism in the human digestive system is extremely important when determining the mechanism of their action in the body. Studies on the concentration of polyphenols in the plasma and urine of mammals suggest that the large part of phenolic compounds contained in the food reaches the small intestine, where they are further metabolized by microbiota. Polyphenols, such as hydroxycinnamic acids, can reach the colon intact and are probably only then broken down by the bacterial microbiota. Simpler compounds resulting from the decomposition of complex polyphenols may have different biological properties. They can have positive effect on health, but also thanks to them modulation of the intestinal microbiota is possible (including bacteria of the genus Lactobacillus) [64].
Metabolism of polyphenols by Lactobacillus spp. bacteria
Among the intestinal microbiota, bacteria of the genus Lactobacillus are characterized by the highest ability to tolerate polyphenolic compounds in the growth environment [65].
Lactobacillus bacteria have many enzymatic abilities that allow them to carry out the transformations described in Table 1.
Phenols, such as hydroxycinnamic acids (caffeic acid, p-coumaric acid, ferulic acid), present in the nutrient matrix affect the growth of the biomass of Lactobacillus bacteria in various ways. First of all, they change the course of metabolic processes such as lactic fermentation. An example of a compound that affects the modulation of LAB microbiota is undoubtedly gallic acid. Low concentrations of gallic acid significantly stimulate the growth and metabolic activity of Lactobacillus bacteria. In the case of high concentrations of gallic acid (above 3 mM), the integrity of the bacterial cell walls is disrupted, resulting in a disturbed pH gradient. Thus, polyphenols can delay carbohydrate metabolism by lactic acid bacteria [69].
The metabolism and degradation of polyphenols by lactic acid bacteria can also provide additional benefits. The phenolic acid reduction process may be an intermediate element in the re-oxidation of the reduced NADH cofactor. Thus, this process provides an energy benefit through NAD+ regeneration. According to research conducted by Silva et al. [72], Lactobacillus plantarum and Lactobacillus collinoides were able to grow in anaerobic conditions in an environment without additional electron acceptors. As a result of the process of reducing 4-vinylphenol to 4-ethylphenol, they increased the availability of NAD+.
Reduction of phenolic acids by LAB can be much more beneficial in strictly heterofermentative species. Species that carry out heterofermentative metabolic processes are characterized by lower energy efficiency than bacteria capable of carrying out homofermentative metabolic processes. Research conducted by Filannino et al. [69] proved that heterofermentative bacteria of the genus Lactobacillus tolerate the presence of phenolic acids in the growth environment in various ways. The minimal value inhibiting the growth of Lactobacillus (MIC) bacteria is a strain feature. It also depends on the group of polyphenols used. The highest ability to tolerate polyphenols in the growth environment was characterized by the Lactobacillus curvatus PE5 strain, whose MIC in the case of caffeic, p-coumaric and ferulic acid exceeded 23.7 mM. Strains belonging to the species Lactobacillus fermentum and Lactobacillus brevis also had high tolerability (23.7 mM concentration) of p-coumaric acid and ferulic acid. Research conducted by Filannino et al. [69] proved that bacteria of the genus Lactobacillus grow relatively poorly in the environment in which the caffeic acid is found (MIC 13.6 mM).
According to research conducted by Gaur et al. [71] heterofermentative bacterial species of the genus Lactobacillus have a much higher ability to degrade hydroxycinnamic acids (using reductases) than homofermentative species. Thus, the degradation of hydroxycinnamic acids is one of the few known processes of polyphenol degradation by LAB. The ability to metabolize hydroxycinnamic acids also depends on the environment from which the strain was isolated. The most frequent reports in the literature describe the ability of bacteria of the species Lactobacillus plantarum to adapt and metabolize polyphenols. Nevertheless, among other plant isolates, this feature is also often attributed to the species Lactobacillus collinoides and Lactobacillus buchneri. Among animal isolates, the species Lactobacillus delbrueckii and Lactobacillus salivarius have the ability to metabolize polyphenols. It is worth remembering that the reduction of phenolic acids by heterofermentative species of lactic acid bacteria causes a decrease in the amount of NADH which, at the same time, ensures higher energy efficiency of the cell.
Enzymatic transformation of polyphenols by Lactobacillus species
Among all species belonging to the genus Lactobacillus, in as many as 68 of them were genes found that are responsible for the ability to synthesize phenolic reductase, while genes responsible for the synthesis of phenolic decarboxylase are characteristic only of bacteria Lactobacillus plantarum. The most widespread species that have the ability to break down phenolic acid include Lactobacillus rossiae, Lactobacillus reuteri, Lactobacillus fermentum and Lactobacillus plantarum. As many as 34 bacterial species of the genus Lactobacillus have genes responsible for the synthesis of more than one type of reductase (e.g. vinylphenol reductase). Genes encoding as many as three different reductases have been found in species such as Lactobacillus plantarum and Lactobacillus pentosus. 19 bacterial species of the genus Lactobacillus have the ability to decarboxylate hydroxycinnamic acids to vinyl derivatives, which in the next stages are transformed into ethyl derivatives [71].
Sometimes, despite the presence of genes encoding enzymes useful in the metabolism of polyphenols (e.g. feruloyl esterase) in the genome, bacteria of the genus Lactobacillus are unable to break them down. An example would be the fact that LABs are not able to metabolize methyl ferulate, methyl caffeine, methyl p-coumarinate or methyl sinapinate [73]. The ability to metabolize polyphenols is therefore a strain trait. Thus, in studies conducted by Gaur et al. [71], it has been proven that Lactobacillus hammesil and Lactobacillus brevis are able to metabolize ferulic and caffeic acid, while the species Lactobacillus reuteri and Lactobacillus kunkeei do not have this ability. Lactobacillus plantarum is believed to have the highest polyphenol metabolism capacity.
Lactobacillus plantarum is undoubtedly the best described bacterial species in terms of the enzymatic transformation of polyphenols. Lactobacillus plantarum has, among other things, the ability to produce benzyl alcohol dehydrogenase, which reversibly oxidizes some aromatic alcohols to aldehydes while reducing NAD+. Lactobacillus plantarum is also capable of hydrolyzing odorless, non-volatile glycosides with the formation of aglycones through the action of glycosidase. Phenolic acid decarboxylase allows Lactobacillus plantarum bacteria to break down p-coumaric acid, caffeic acid and ferulic acid. In addition, Lactobacillus plantarum bacteria are able to hydrolyse ester bonds in tannins and gallic acid esters due to their ability to produce tannin acyl hydrolase. Among other enzymes, the synthesis of which Lactobacillus bacteria are capable of, include esterases, reductases, and decarboxylases [74].
Examples of metabolic transformations using enzymes synthesized by Lactobacillus plantarum are presented in Table 2.
The ability of Lactobacillus plantarum to metabolize polyphenols may be due to the fact that this species is commonly found in products of plant origin. Plant fermentation requires Lactobacillus plantarum to have enzymatic capabilities that ensure the ability to ferment phenolic compounds. Thus, the use of Lactobacillus plantarum as a starter culture in the production of fermented foods carries some risk of organoleptic changes in the finished product. Due to the fact that these bacteria are capable of metabolizing hydroxycinnamic acids with the simultaneous release of vinyl and ethyl derivatives (which can react with anthocyanins and 3-deoxyanthocyanins to form pyrananthocyanins and 3-deoxypiroanthocyanins), there may be changes in the aroma of fermented food. On the other hand, by releasing hydroxycinnamates, Lactobacillus plantarum helps to increase a product’s health value. Hydrocinnamates are compounds with antioxidant, anti-inflammatory and antimicrobial activity [71, 73]. Lactobacillus plantarum, as the only one in the genus Lactobacillus, also has the ability to synthesize the enzyme tannase. Owing to this, Lactobacillus plantarum is able to metabolize tannins (tannic acid) to gallic acid and glucose. In subsequent stages, bacteria convert gallic acid to pyrogallol [78]. Therefore, the enzymatic capabilities of bacteria of the genus Lactobacillus may be of particular importance in the selection of starter cultures in processes based on food fermentation. The new trend in functional food is the removal of antinutritional factors (ANF) from natural products, among which some polyphenols can be identified. Tannins that can be found in some foods are considered undesirable compounds (due to their ability to inhibit digestive enzymes). In studies conducted by Saez et al. [79] it was demonstrated that two strains of Lactobacillus plantarum had the ability to synthesize decarboxylases capable of breaking down tannins and gallic acid.
Prebiotic activity of polyphenols
Apart from their ability to limit the development of microbiota considered as pathogenic, some polyphenols (and their metabolites) are able to stimulate the growth of commensal bacteria populating the human intestinal system [80]. Some polyphenols are able to inhibit the multiplication of microbiota exhibiting a probiotic potential; in most cases, they demonstrate prebiotic characteristics. According to the research conducted by Attri, Goel [81], polyphenols derived from sea buckthorn, during 10-day supplementation, may increase the multiplication of lactic acid bacteria 1.18-fold. Despite the initial nearly 20%, inhibition of bacteria multiplication, after a 48-h incubation period, lactic bacteria adapt to environmental conditions. The biomass increases its size up to approximately 10 logarithmic units (LU). A similar effect was also observed in tests conducted by Dolara et al. [82]. Feeding rats with red wine powder resulted in a considerable increase in the number of Lactobacillus genus bacteria in animal faeces. On the other hand, the tests conducted by Tabasco et al. [10] show that polyphenols (including catechin, epicatechin and flavan-3-ol), in concentrations of 0.25–1.0 mg/ml, effectively become growth stimulators for strains of LAB. More interestingly, when these polyphenols are used as individual compounds, they sometimes contribute to the inhibition of the growth of such bacteria as Lactobacillus plantarum, Lactobacillus casei and Lactobacillus bulgaricus.
Tests conducted on groups of mammals (rats, pigs and humans) and birds (chickens) proved that polyphenols isolated from grape pip extracts [83], blackcurrants [84], and wine [82] demonstrate a prebiotic potential in relation to Lactobacillus genus bacteria. Simultaneously, they contribute to inhibition of the multiplication of the Enterobacteriaceae family and Bacteroides and Clostridium genus bacteria. Their ability to demonstrate a prebiotic potential enables polyphenols to influence the modulation of intestinal microbiota [85].
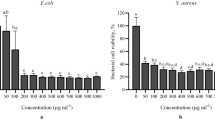
Phenolic compounds (concentration: 10% v/v–2.9 mg/ml) contained in sunflower flour contribute significantly to enhancing the metabolic capabilities of lactic acid bacteria and producing significantly larger amounts of lactic acid. This results from the fact that polyphenols contained in sunflower seeds ensure the higher development potential of the following bacteria: Bifidobacterium animalis subsp. lactis, Lactobacillus plantarum and Lactobacillus gasseri. Phenolic acids from sunflower flour are an additional source of carbon [86] in this environment. Undoubtedly, another polyphenolic compound influencing the development of the LAB is chlorogenic acid. It must be stressed that metabolites generated during interactions between polyphenols and intestinal microbiota may be a factor conditioning its multiplication. This effect can be bidirectional, i.e. they can be either activators or inhibitors of intestinal microbiota growth. Not all lactic acid bacteria are able to develop in the presence of polyphenolic compounds. According to the research conducted by Stead [87], hydroxycinnamic, caffeic, coumaric and ferulic acids inhibit the development of Lactobacillus brevis bacteria, at a concentration of 0.5 mg/ml. However, the tests conducted by Parkar et al. [88] proved that chlorogenic and caffeic acids, at the concentration of 0.25 mg/ml, inhibit the growth of Lactobacillus rhamnosus bacteria. Generally, lactic acid bacteria demonstrate much higher resistance to polyphenols, in comparison to pathogenic microbiota. The decrease in the number of lactic acid bacteria (Lactobacillus plantarum, Lactobacillus gasseri and Pediococcus pentosaceus), during a 24-h incubation period, only takes place after a 10–20 mg/ml concentration of polyphenols is administered. For example, a 24-h period of incubation of such pathogenic bacteria as Escherichia coli, Staphylococcus aureus and Salmonella enterica in the presence of chlorogenic acid, at 100 times lower concentration (0.1–0.2 mg/ml), results in significant inhibition of their growth [88, 89]. Polyphenol-rich melon juice (at pH 6.7) demonstrates bactericidal properties in relation to the probiotic strain of Lactobacillus rhamnosus only at a concentration of 5 mg/ml. On the other hand, an addition of melon juice at a concentration of 10 mg/ml to a cultivation base does not inhibit multiplication of such lactic acid bacteria as Lactobacillus delbrueckii subsp. bulgaricus, Lactobacillus paracasei subsp. paracasei and Lactococcus lactis subsp. cremoris [63].
The positive effect of the prebiotic potential of compounds resulting from polyphenol metabolism results in the fact that the level of multiplication of probiotic intestinal microbiota is significantly higher. This, in turn, ensures better protection against stomach and intestinal disorders and additional protection against pathogenic microorganisms [90, 91]. In research on polyphenols present in pomace extracts from pseudo-fruit Rosa rugosa Thunb., Piekarska-Radzik et al. [92] proved that depending on the concentration used, the extracts can act as a factor stimulating the growth of Lactobacillus bacteria. Extracts from Rosa rugosa Thunb. (rich in polyphenols such as ellagitannins, free ellagic acid, flavonols, procyanidins and free catechins) are characterized by a total content of polyphenols at the level of 8.8–33.2 g/100 g dry matter. Thus, the addition of extracts with polyphenols at 0.156 mg/ml concentration to the bacteria culture of the genus Lactobacillus is the main factor determining of biomass growth. In case of the Lactobacillus acidophilus ŁOCK 0928 strain, the water–ethanol extract polyphenols increased biomass growth by almost 15% compared to the control culture. With Lactobacillus brevis ŁOCK 0944 bacteria, a higher degree of biomass multiplication (7–10%) was observed in cultures with the addition of purified water–ethanol extract and crude water–acetone polyphenols. The authors also presented the results of studies showing that polyphenols at a concentration of 0.156 mg/ml (purified water–ethanol extract) added to the Lactobacillus casei ŁOCK 0979 are a factor stimulating biomass growth (by about 16% compared to the control culture). Thus, Piekarska-Radzik et al. [92], proved that bacteria of the genus Lactobacillus have the ability to tolerate polyphenols in the environment. They also emphasize the prebiotic potential of polyphenols contained in pseudo-fruit of Rosa rugosa Thunb.
In studies conducted by Filannino et al. [70], 54 strains of bacteria from the genus Lactobacillus constituting the intestinal microbiota of honeybee (Apis mellifera L.) were isolated—52 strains from the species Lactobacillus kunkeei, and one strain each of Lactobacillus plantarum and Lactobacillus fermentum. Further analysis showed that only 11 strains among the Lactobacillus kunkeei species could be classified as FLAB (fructophilic lactic acid bacteria). Interestingly, fructophilic lactation bacteria placed in the digestive tract of a honeybee was able to grow in an environment containing polyphenols such as p-coumaric acid, caffeic acid, gallic acid and syringic acid. The minimum value inhibiting bacterial growth was 22.7 mmol/L. The addition of Lactobacillus kunkeei to the above-mentioned polyphenols in the 1 mmol/L culture to the culture did not significantly affect the biomass increase. Nevertheless, Filannino et al. [70] observed that FLAB has the ability to consume secondary metabolites of plants. Lactobacillus kunkeei degraded p-coumaric acid in 57–78%, caffeic acid 43–50%, syringic acid 40–53% and gallic acid 42–51%.
In the literature, there is also other evidence confirming the prebiotic properties of plant extracts rich in polyphenols. Examples of plants characterized by a high content of polyphenolic compounds that favourably affect the growth of Lactobacillus bacteria are presented in Table 3.
Sometimes, incubation of Lactobacillus bacteria (especially those with probiotic potential) can change the specific strain characteristics. Among these, the production of short-chain fatty acids (SCFA) can be distinguished. Zhang et al. [98] proved that polyphenols abundantly found in oolong tea (epigallocatechin gallate, galactocatechin gallate) not only significantly affected the growth of Lactobacillus bacteria. The addition to the tea-based extract culture resulted in SCFA synthesis at a much higher level, particularly formic, acetic, propionic and butyric acid. Interestingly, during 24-h incubation, the synthesis of lactic acid (which is the base for the production of other SCFAs) by Lactobacillus spp. in the control sample occurred much more slowly than in cultures with the addition of polyphenols.
Influence of polyphenols on Lactobacillus spp. gene regulation
The interaction between polyphenols supplied with food and the microbiota inhabiting the human digestive tract is not limited to changes in the level of biomass multiplication. Phenolic compounds are able to cause changes in gene expression in bacteria belonging to the Lactobacillus species.
In studies on the interaction between polyphenols and intestinal microbiota by Reverón et al. [64], a strain of Lactobacillus plantarum was used. After a 10-min incubation of bacteria together with p-coumaric acid at a concentration of 1.5 mM, significant differences in gene expression (9%) were observed. 280 transcripts changed (increased expression in 144 genes; reduced expression in 136 genes). The largest group of genes whose expression was changed was responsible for amino acid transport and metabolism, translation and cell wall and membrane synthesis. The shock caused by the presence of polyphenols in the environment caused that Lactobacillus plantarum expressed the genes coding proteases or heat shock proteins at a higher level. In the case of the Lactobacillus plantarum species, similar results were achieved in studies conducted by Esteban-Torres et al. [73]. Under the influence of olive oil rich in polyphenols such as hydroxytyrosol (9.31 µg/g) and p-coumaric acid (0.14 µg/g), 230 transcripts changed (increased expression in 123 genes; reduced expression in 107 genes). In turn, research by Santamaria et al. [99] regarding changes in gene expression in the genome of Lactobacillus plantarum under the influence of oleuropeins (concentration 15 mM) proved that after a 10-min incubation in the bacterial genome there was a change in the expression of as many as 358 genes (increased expression of 155 genes; reduced expression of 203 genes).
There is evidence in the literature that many polyphenolic compounds have the ability to alter the gene expression of Lactobacillus bacteria. Changes were observed after the addition of, among others, polyphenols such as jasmonic acid, jasmonate, gallic acid, salicylic acid and resveratrol to culture media [99].
The addition of polyphenols can contribute to changes in the expression of genes responsible for the synthesis of specific enzymes, genes encoding proteins (including those responsible for transport), and the induction of many metabolic processes [64, 73, 77, 99, 100].
Polyphenols often found in food (such as hydroxycinnamic acids) in bacteria of the genus Lactobacillus can cause increased expression of the genes responsible for the transport of purines, oligopeptides (ATP-dependent transport), branched chain amino acids, methionine, Na+ ions (indirect effect on homeostasis regulation), dihydroxyacetone, saccharides (mannose, mannitol, cellobiose, fructose). At the same time, they contribute to reduced transport of pyrimidines, NH4+ ions, N-acyl-glucosamine or electrons (disorders in respiratory metabolism) [64, 73, 77, 99, 100].
Polyphenols are also responsible for increased expression of Lactobacillus spp. genes responsible for biosynthesis of chorismate, asparagine, methionine, cell wall surface proteins (increased synthesis of peptidoglycan; strengthening the cell wall; increase survival in the gastrointestinal tract), oxidative shock proteins (antioxidant oxidation protein), proteins involved in cell proliferation, polysaccharides (extracellular), d-lactate (the basic component of peptidoglycan); protein acetylation, lactate racemization, transpeptization; tRNA accumulation. At the same time, under the influence of polyphenols, the expression of genes responsible for biosynthesis is reduced: purines, capsular polysaccharides, serine, glutamine, cysteine, proteins constituting the autoaggregation factor, proteins involved in the FAB pathway, fatty acids, transcription factor constituting a global repressor of nitrogen metabolism; cell assimilation of nitrogen (altered production of glutamine and ammonium ions) [64, 73, 77, 99, 100].
Polyphenols also affect the expression of genes responsible for the synthesis of many specific enzymes. As a result of the action of polyphenols, there is an increased expression of genes responsible for the synthesis of enzymes such as transketolase, phosphoglycerate mutase, phosphoketolase, glutathione reductase, methionine sulfoxide reductase, oxidoreductase [NAD-dependent (P)], quinoxone oxidoreductase, quinoxone oxidase pyruvate metabolism), glycosidase transferase, acetyl CoA synthase, glycerol phosphate dehydrogenase, glyceryl kinase, d-lactate dehydrogenase, peptidoglycan hydrolase), glucosamine-6-phosphate deaminase, glucosamine-6-phosphate isomerase. On the other hand, the expression of genes responsible for the synthesis of phosphoglycerate dehydrogenase, nitrate reductase, peptide glycan hydrolase, N-glucosamine phosphotransferase, galactosamine phosphotransferase, acetate kinase, methionyl-tRNA formyltransferase, and cell wall transglycosidase is reduced [64, 73, 77, 99, 100].
Summary
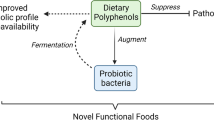
The trend of combining both probiotic bacteria and polyphenols in food has caused that more products on the food market are appearing in the “functional food” category. Attempts to identify mechanisms connecting polyphenols and probiotic bacterial species of the Lactobacillus genus have led to the conclusion that the interaction between both groups can significantly affect the survival time of the bacteria in a product. Such factors as the common presence of lactic acid bacteria in the environment and the high content of polyphenols in plant-derived products have facilitated the development of unique interactions between them. From the point of view of higher organisms, the activity of intestinal flora, in particular Lactobacillus genus bacteria, results in an increase in the antioxidation potential of polyphenols. Due to the specificity of the metabolism of plant compounds contained in food, lactic acid bacteria are able to assist the host organism to assimilate compounds demonstrating antioxidative activity, frequently gaining access to substances demonstrating a prebiotic potential. Moreover, in the presence of polyphenols, lactic acid bacteria ensure higher concentrations of lactic acid, which simultaneously stabilizes phytocompounds. A combination of correctly selected polyphenols and LAB bacteria in the food matrix makes it possible to produce food demonstrating a high functional potential. Such research conforms to the current worldwide policy of searching for food that demonstrates high nutritional potential and ensures microbiological safety.
It is worth emphasizing how important it is to learn about the mechanisms linking polyphenols with microorganisms. The interactions of compounds of plant origin present in food, with the intestinal microbiota are extremely important. Changing gene expression in both microorganisms with prebiotic potential and pathogenic (food contaminating) microbiota directly affects human health. By adding appropriately selected polyphenols to food, it is possible to influence gene expression responsible for virulence (i.e. Escherichia coli), protecting potential consumers against infections. Thus, it is worth noting that it is possible to design modern food protection systems against undesirable microbiota that contaminate food.
Polyphenols delivered with the diet to the human gastrointestinal tract may behave either as antioxidants or pro-oxidants. By appropriate regulation of the pH (especially in food products), it is possible to create a favourable pro-oxidative environment in which polyphenols turn out to be bactericidal against pathogenic bacteria. On the other hand, from the point of view of bacteria belonging to the genus Lactobacillus, hydrogen peroxide generated in the process of self-oxidation of polyphenols would introduce a mild degree of oxidative stress (which, in consequence, may lead to increased expression of genes encoding catalase, superoxide dismutase—higher survivability). Interestingly, bacteria of the genus Lactobacillus also have the ability to produce H2O2 (a by-product of the process of aerobic cellular respiration or oxidation of hydrocarbons), which limits the development of undesirable intestinal microbiota and promotes the regeneration of the intestinal epithelium. The combination of polyphenols and Lactobacillus spp. bacteria may therefore have a significant impact on maintaining homeostasis in the human gastrointestinal tract.
References
Rasouli H, Farzaei MH, Khodarahmi R (2017) Polyphenols and their benefits: a review. Int J Food Prop 20(sup2):1700–1741. https://doi.org/10.1080/10942912.2017.1354017
Lutomski J, Mścisz A (2003) The preventive importance of polyphenolic compounds contained in grapes. Post Fitoter 1:6–10
Manach C, Scalbert A, Morand C, Remesy C, Jimenez L (2004) Polyphenols: food sources and bioavailability. Am J Clin Nutr 79(5):727–747. https://doi.org/10.1093/ajcn/79.5.727
Kosiorek A (2013) Basics for the use of plant polyphenols as nutraceuticals with antiplatelet properties. Post Fitoter 14(2):108–117
Gherini E (2011) Związki polifenolowe w owocach i warzywach. Med Rodz 14(4):111–115
Pandey KB, Rizvi SI (2009) Plant polyphenols as dietary antioxidants in human health and disease. Oxid Med Cell Longev 2(5):270–278. https://doi.org/10.4161/oxim.2.5.9498
Li R, Fei P, Man CX, Lou BB, Niu JT, Feng J, Sun LH, Li MY, Jiang YJ (2016) Tea polyphenols inactivate Cronobacter sakazakii isolated from powdered infant formula. J Dairy Sci 99(2):1019–1028. https://doi.org/10.3168/jds.2015-10039
Yi S, Wang W, Bai F, Zhu J, Li J, Li X, Xu Y, Sun T, He Y (2014) Antimicrobial effect and membrane-active mechanism of tea polyphenols against Serratia marcescens. World J Microb Biot 30(2):451–460. https://doi.org/10.1007/s11274-013-1464-4
Zhang H, Zhou W, Zhang W, Yang A, Liu Y, Jiang Y, Huang S, Su J (2014) Inhibitory effects of citral, cinnamaldehyde, and tea polyphenols on mixed biofilm formation by foodborne Staphylococcus aureus and Salmonella enteritidis. J Food Prot 77(6):927–933. https://doi.org/10.4315/0362-028X.JFP-13-497
Tabasco R, Sanchez-Patan F, Monagas M, Bartolome B, Victoria Moreno-Arribas M, Pelaez C, Requena T (2011) Effect of grape polyphenols on lactic acid bacteria and bifidobacteria growth: resistance and metabolism. Food Microbiol 28(7):1345–1352. https://doi.org/10.1016/j.fm.2011.06.005
Piekut J (2017) Evaluation of selected extracts of spice plants in terms of their antimicrobial properties and the content of phenolic acids. Zeszyty Problemowe Postępów Nauk Rolniczych 588:103–111. https://doi.org/10.22630/ZPPNR.2017.588.10
Othman L, Sleiman A, Abdel-Massih RM (2019) Antimicrobial activity of polyphenols and alkaloids in Middle Eastern plants. Front Mikrobiol 10:911
Tepe B, Sokmen M, Akpulat HA, Sokmen A (2005) In vitro antioxidant activities of the methanol extracts of five Allium species from Turkey. Food Chem 92(1):89–92. https://doi.org/10.1016/j.foodchem.2004.07.016
Mai-Prochnow A, Clauson M, Hong J, Murphy AB (2016) Gram positive and Gram negative bacteria differ in their sensitivity to cold plasma. Sci Rep 6(1):38610. https://doi.org/10.1038/srep38610
Silhavy TJ, Kahne D, Walker S (2010) The bacterial cell envelope. Cold Spring Harb Perspect Biol 2(5):a000414. https://doi.org/10.1101/cshperspect.a000414
Yu X, Chu S, Hagerman AE, Lorigan GA (2011) Probing the interaction of polyphenols with lipid bilayers by solid-state NMR spectroscopy. J Agric Food Chem 59(12):6783–6789. https://doi.org/10.1021/jf200200h
Borges A, Ferreira C, Saavedra MJ, Simões M (2013) Antibacterial activity and mode of action of ferulic and gallic acids against pathogenic bacteria. Microb Drug Resist 19(4):256–265. https://doi.org/10.1089/mdr.2012.0244
Ultee A, Bennik MH, Moezelaar R (2002) The phenolic hydroxyl group of carvacrol is essential for action against the food-borne pathogen Bacillus cereus. Appl Environ Microbiol 68(4):1561–1568
Gao H, Cheng N, Zhou J, Wang B, Deng J, Cao W (2014) Antioxidant activities and phenolic compounds of date plum persimmon (Diospyros lotus L.) fruits. Int J Food Sci Technol 51(5):950–956. https://doi.org/10.1007/s13197-011-0591-x
Baranowska M, Bartoszek A (2016) Antioxidant and antimicrobial properties of bioactive phytochemicals from cranberry. Post Hig Med Dosw (Online) 70:1460–1468. https://doi.org/10.5604/17322693.1227896
Yamanaka A, Kimizuka R, Kato T, Okuda K (2004) Inhibitory effects of cranberry juice on attachment of oral streptococci and biofilm formation. Oral Microbiol Immunol 19(3):150–154. https://doi.org/10.1111/j.0902-0055.2004.00130.x
Hisano M, Bruschini H, Nicodemo AC, Srougi M (2012) Cranberries and lower urinary tract infection prevention. Clinics (Sao Paulo) 67(6):661–668
Rahman T, Hosen I, Islam MMT, Shekhar HU (2012) Oxidative stress and human health. ABB 3(7A):997–1019
Chen M, Zhao Z, Meng H, Yu S (2017) The antibiotic activity and mechanisms of sugar beet (Beta vulgaris) molasses polyphenols against selected food-borne pathogens. LWT Food Sci Technol 82:354–360. https://doi.org/10.1016/j.lwt.2017.04.063
Zhao Y, Chen M, Zhao Z, Yu S (2015) The antibiotic activity and mechanisms of sugarcane (Saccharum officinarum L.) bagasse extract against food-borne pathogens. Food Chem 185:112–118. https://doi.org/10.1016/j.foodchem.2015.03.120
Wang C, Chang T, Yang H, Cui M (2015) Antibacterial mechanism of lactic acid on physiological and morphological properties of Salmonella Enteritidis, Escherichia coli and Listeria monocytogenes. Food Control 47:231–236. https://doi.org/10.1016/j.foodcont.2014.06.034
Zeng X, Tang W, Ye G, Ouyang T, Tian L, Ni Y, Li P (2010) Studies on disinfection mechanism of electrolyzed oxidizing water on E. coli and Staphylococcus aureus. J Food Sci Technol 75(5):M253–260. https://doi.org/10.1111/j.1750-3841.2010.01649.x
Ricke SC (2003) Perspectives on the use of organic acids and short chain fatty acids as antimicrobials. Poult Sci 82(4):632–639. https://doi.org/10.1093/ps/82.4.632
Fei P, Ali MA, Gong S, Sun Q, Bi X, Liu S, Guo L (2018) Antimicrobial activity and mechanism of action of olive oil polyphenols extract against Cronobacter sakazakii. Food Control 94:289–294. https://doi.org/10.1016/j.foodcont.2018.07.022
Halliwell B (1990) How to characterize a biological antioxidant. Free Radic Res Commun 9(1):1–32. https://doi.org/10.3109/10715769009148569
Grzesik M, Bartosz G, Stefaniuk I, Pichla M, Namiesnik J, Sadowska-Bartosz I (2019) Dietary antioxidants as a source of hydrogen peroxide. Food Chem 278:692–699. https://doi.org/10.1016/j.foodchem.2018.11.109
Macvanin M, Hughes D (2010) Assays of sensitivity of antibiotic-resistant bacteria to hydrogen peroxide and measurement of catalase activity. Methods Mol Biol 642:95–103. https://doi.org/10.1007/978-1-60327-279-7_7
Daglia M (2012) Polyphenols as antimicrobial agents. Curr Opin Biotechnol 23(2):174–181. https://doi.org/10.1016/j.copbio.2011.08.007
Halliwell B (2008) Are polyphenols antioxidants or pro-oxidants? What do we learn from cell culture and in vivo studies? Arch Biochem Biophys 476(2):107–112. https://doi.org/10.1016/j.abb.2008.01.028
Liu X, Li J, Wang Y, Li T, Zhao J, Zhang C (2013) Green tea polyphenols function as prooxidants to inhibit Pseudomonas aeruginosa and induce the expression of oxidative stress-related genes. Folia Microbiol (Praha) 58(3):211–217. https://doi.org/10.1007/s12223-012-0198-2
Dai J, Mumper RJ (2010) Plant phenolics: extraction, analysis and their antioxidant and anticancer properties. Molecules 15(10):7313–7352. https://doi.org/10.3390/molecules15107313
Bucekova M, Buriova M, Pekarik L, Majtan V, Majtan J (2018) Phytochemicals-mediated production of hydrogen peroxide is crucial for high antibacterial activity of honeydew honey. Sci Rep 8(1):9061. https://doi.org/10.1038/s41598-018-27449-3
de Graft-Johnson J, Nowak D (2016) Effect of selected plant phenolics on Fe(2+)-EDTA-H(2)O(2) system mediated deoxyribose oxidation: molecular structure-derived relationships of anti- and pro-oxidant actions. Molecules. https://doi.org/10.3390/molecules22010059
Brudzynski K, Abubaker K, Miotto D (2012) Unraveling a mechanism of honey antibacterial action: polyphenol/H(2)O(2)-induced oxidative effect on bacterial cell growth and on DNA degradation. Food Chem 133(2):329–336. https://doi.org/10.1016/j.foodchem.2012.01.035
Taleb H, Maddocks SE, Morris RK, Kanekanian AD (2016) The antibacterial activity of date syrup polyphenols against S. aureus and E. coli. Front Microbiol 7:198. https://doi.org/10.3389/fmicb.2016.00198
Sakihama Y, Cohen MF, Grace SC, Yamasaki H (2002) Plant phenolic antioxidant and prooxidant activities: phenolics-induced oxidative damage mediated by metals in plants. Toxicology 177(1):67–80. https://doi.org/10.1016/s0300-483x(02)00196-8
Yordi E, Molina Pérez E, Matos MJ, Uriarte E (2012) Antioxidant and pro-oxidant effects of polyphenolic compounds and structure-activity relationship evidence. Nutrition. https://doi.org/10.5772/29471
Almajano MP, Carbo R, Delgado ME, Gordon MH (2007) Effect of pH on the antimicrobial activity and oxidative stability of oil-in-water emulsions containing caffeic acid. J Food Sci 72(5):C258–263. https://doi.org/10.1111/j.1750-3841.2007.00387.x
Akagawa M, Shigemitsu T, Suyama K (2003) Production of hydrogen peroxide by polyphenols and polyphenol-rich beverages under quasi-physiological conditions. Biosci Biotechnol Biochem 67(12):2632–2640. https://doi.org/10.1271/bbb.67.2632
Long LH, Hoi A, Halliwell B (2010) Instability of, and generation of hydrogen peroxide by, phenolic compounds in cell culture media. Arch Biochem Biophys 501(1):162–169. https://doi.org/10.1016/j.abb.2010.06.012
Poli JP, Guinoiseau E, Luciani A, Yang Y, Battesti MJ, Paolini J, Costa J, Quilichini Y, Berti L, Lorenzi V (2018) Key role of hydrogen peroxide in antimicrobial activity of spring, Honeydew maquis and chestnut grove Corsican honeys on Pseudomonas aeruginosa DNA. Lett Appl Microbiol 66(5):427–433. https://doi.org/10.1111/lam.12868
Lluis L, Munoz M, Nogues MR, Sanchez-Martos V, Romeu M, Giralt M, Valls J, Sola R (2011) Toxicology evaluation of a procyanidin-rich extract from grape skins and seeds. Food Chem Toxicol 49(6):1450–1454. https://doi.org/10.1016/j.fct.2011.03.042
Gomes FMS, da Cunha XJ, Dos Santos JFS, de Matos Y, Tintino SR, de Freitas TS, Coutinho HDM (2018) Evaluation of antibacterial and modifying action of catechin antibiotics in resistant strains. Microb Pathog 115:175–178. https://doi.org/10.1016/j.micpath.2017.12.058
Lambert JD, Kwon SJ, Hong J, Yang CS (2007) Salivary hydrogen peroxide produced by holding or chewing green tea in the oral cavity. Free Radic Res 41(7):850–853. https://doi.org/10.1080/10715760601091659
Hiramoto K, Kida T, Kikugawa K (2002) Increased urinary hydrogen peroxide levels caused by coffee drinking. Biol Pharm Bull 25(11):1467–1471. https://doi.org/10.1248/bpb.25.1467
Smith AH, Zoetendal E, Mackie RI (2005) Bacterial mechanisms to overcome inhibitory effects of dietary tannins. Microb Eco 50(2):197–205. https://doi.org/10.1007/s00248-004-0180-x
Kutschera M, Engst W, Blaut M, Braune A (2011) Isolation of catechin-converting human intestinal bacteria. J Appl Microbiol 111(1):165–175. https://doi.org/10.1111/j.1365-2672.2011.05025.x
Curiel JA, Rodríguez H, Landete JM, de Las Rivas B, Muñoz R (2010) Ability of Lactobacillus brevis strains to degrade food phenolic acids. Food Chem 120(1):225–229. https://doi.org/10.1016/j.foodchem.2009.10.012
Gross G, Jacobs DM, Peters S, Possemiers S, van Duynhoven J, Vaughan EE, van de Wiele T (2010) In vitro bioconversion of polyphenols from black tea and red wine/grape juice by human intestinal microbiota displays strong interindividual variability. J Agric Food Chem 58(18):10236–10246. https://doi.org/10.1021/jf101475m
Kwaw E, Ma Y, Tchabo W, Apaliya MT, Wu M, Sackey AS, Xiao L, Tahir HE (2018) Effect of lactobacillus strains on phenolic profile, color attributes and antioxidant activities of lactic-acid-fermented mulberry juice. Food Chem 250:148–154. https://doi.org/10.1016/j.foodchem.2018.01.009
Pérez-Gregorio MR, Regueiro J, Alonso-González E, Pastrana-Castro LM, Simal-Gándara J (2011) Influence of alcoholic fermentation process on antioxidant activity and phenolic levels from mulberries (Morus nigra L.). LWT Food Sci Tech 44(8):1793–1801. https://doi.org/10.1016/j.lwt.2011.03.007
Hur SJ, Lee SY, Kim YC, Choi I, Kim GB (2014) Effect of fermentation on the antioxidant activity in plant-based foods. Food Chem 160:346–356. https://doi.org/10.1016/j.foodchem.2014.03.112
Pedrosa MM, Muzquiz M, García-Vallejo C, Burbano C, Cuadrado C, Ayet G, Robredo LM (2000) Determination of caffeic and chlorogenic acids and their derivatives in different sunflower seeds. J Sci Food Agric 80(4):459–464. https://doi.org/10.1002/(SICI)1097-0010(200003)80:4<459:AID-JSFA549>3.0.CO;2-O
Weisz GM, Kammerer DR, Carle R (2009) Identification and quantification of phenolic compounds from sunflower (Helianthus annuus L.) kernels and shells by HPLC-DAD/ESI-MSn. Food Chem 115(2):758–765. https://doi.org/10.1016/j.foodchem.2008.12.074
Filannino P, Bai Y, Di Cagno R, Gobbetti M, Ganzle MG (2015) Metabolism of phenolic compounds by Lactobacillus spp. during fermentation of cherry juice and broccoli puree. Food Microbiol 46:272–279. https://doi.org/10.1016/j.fm.2014.08.018
Sanchez-Maldonado AF, Schieber A, Ganzle MG (2011) Structure-function relationships of the antibacterial activity of phenolic acids and their metabolism by lactic acid bacteria. J Appl Bacteriol 111(5):1176–1184. https://doi.org/10.1111/j.1365-2672.2011.05141.x
Svensson L, Sekwati-Monang B, Lutz DL, Schieber A, Ganzle MG (2010) Phenolic acids and flavonoids in nonfermented and fermented red sorghum (Sorghum bicolor (L.) Moench). J Agric Food Chem 58(16):9214–9220. https://doi.org/10.1021/jf101504v
Rúa J, López-Rodríguez I, Sanz J, García-Fernández MC, del Valle MP, García-Armesto MR (2018) Improving functional properties of “Piel de Sapo” melon juice by addition of a Lippia citriodora natural extract and probiotic-type lactic acid bacteria. LWT Food Sci Technol 96:75–81. https://doi.org/10.1016/j.lwt.2018.05.028
Reverón I, de Las Rivas B, Muñoz R, de LópezFelipe F (2012) Genome-wide transcriptomic responses of a human isolate of Lactobacillus plantarum exposed to p-coumaric acid stress. Mol Nutr Food Res 56(12):1848–1859. https://doi.org/10.1002/mnfr.201200384
Reveron I, de las Rivas B, Matesanz R, Muñoz R, de LópezFelipe F (2015) Molecular adaptation of Lactobacillus plantarum WCFS1 to gallic acid revealed by genome-scale transcriptomic signature and physiological analysis. Microb Cell Fact 14:160. https://doi.org/10.1186/s12934-015-0345-y
Alberto MR, Gomez-Cordoves C, Manca de Nadra MC (2004) Metabolism of gallic acid and catechin by Lactobacillus hilgardii from wine. J Agric Food Chem 52(21):6465–6469. https://doi.org/10.1021/jf049239f
Bloem A, Bertrand A, Lonvaud-Funel A, de Revel G (2007) Vanillin production from simple phenols by wine-associated lactic acid bacteria. Lett Appl Microbiol 44(1):62–67. https://doi.org/10.1111/j.1472-765X.2006.02037.x
Enol A, Couto J, Campos F, Figueiredo A, Hogg T, Researcher S (2006) Ability of lactic acid bacteria to produce volatile phenols. Am J Enol Viticult 57:166–171
Filannino P, Gobbetti M, De Angelis M, Di Cagno R (2014) Hydroxycinnamic acids used as external acceptors of electrons: an energetic advantage for strictly heterofermentative lactic acid bacteria. Appl Environ Microbiol 80(24):7574–7582. https://doi.org/10.1128/AEM.02413-14
Filannino P, Di Cagno R, Addante R, Pontonio E, Gobbetti M (2016) Metabolism of fructophilic lactic acid bacteria isolated from the Apis mellifera L. bee gut: phenolic acids as external electron acceptors. Appl Environ Microbiol 82(23):6899–6911. https://doi.org/10.1128/AEM.02194-16
Gaur G, Oh JH, Filannino P, Gobbetti M, van Pijkeren JP, Ganzle MG (2020) Genetic determinants of hydroxycinnamic acid metabolism in heterofermentative lactobacilli. Appl Environ Microbiol. https://doi.org/10.1128/AEM.02461-19
Silva I, Campos FM, Hogg T, Couto JA (2011) Factors influencing the production of volatile phenols by wine lactic acid bacteria. Int J Food Microbiol 145(2–3):471–475. https://doi.org/10.1016/j.ijfoodmicro.2011.01.029
Esteban-Torres M, Landete JM, Reveron I, Santamaria L, de Las Rivas B, Muñoz R (2015) A Lactobacillus plantarum esterase active on a broad range of phenolic esters. Appl Environ Microbiol 81(9):3235–3242. https://doi.org/10.1128/AEM.00323-15
Rodriguez H, Curiel JA, Landete JM, de Las Rivas B, de LopezFelipe F, Gomez-Cordoves C, Mancheno JM, Muñoz R (2009) Food phenolics and lactic acid bacteria. Int J Food Microbiol 132(2–3):79–90. https://doi.org/10.1016/j.ijfoodmicro.2009.03.025
Gury J, Barthelmebs L, Tran NP, Divies C, Cavin JF (2004) Cloning, deletion, and characterization of PadR, the transcriptional repressor of the phenolic acid decarboxylase-encoding padA gene of Lactobacillus plantarum. Appl Environ Microbiol 70(4):2146–2153. https://doi.org/10.1128/aem.70.4.2146-2153.2004
Landete JM, Curiel JA, Rodríguez H, de Las Rivas B, Muñoz R (2008) Study of the inhibitory activity of phenolic compounds found in olive products and their degradation by Lactobacillus plantarum strains. Food Chem 107(1):320–326. https://doi.org/10.1016/j.foodchem.2007.08.043
Santamaria L, Reveron I, de Felipe FL, de Las Rivas B, Munoz R (2018) Ethylphenol formation by Lactobacillus plantarum: identification of the enzyme involved in the reduction of vinylphenols. Appl Environ Microbiol. https://doi.org/10.1128/AEM.01064-18
Reveron I, Jimenez N, Curiel JA, Penas E, Lopez de Felipe F, de Las Rivas B, Munoz R (2017) Differential gene expression by Lactobacillus plantarum WCFS1 in response to phenolic compounds reveals new genes involved in tannin degradation. Appl Environ Microbiol. https://doi.org/10.1128/AEM.03387-16
Saez GD, Flomenbaum L, Zarate G (2018) Lactic acid bacteria from argentinean fermented foods: isolation and characterization for their potential use as starters for fermentation of vegetables. Food Technol Biotechnol 56(3):398–410. https://doi.org/10.17113/ftb.56.03.18.5631
Cueva C, Moreno-Arribas MV, Martin-Alvarez PJ, Bills G, Vicente MF, Basilio A, Rivas CL, Requena T, Rodriguez JM, Bartolome B (2010) Antimicrobial activity of phenolic acids against commensal, probiotic and pathogenic bacteria. Res Microbiol 161(5):372–382. https://doi.org/10.1016/j.resmic.2010.04.006
Attri S, Goel G (2018) Influence of polyphenol rich seabuckthorn berries juice on release of polyphenols and colonic microbiota on exposure to simulated human digestion model. Food Res Int 111:314–323. https://doi.org/10.1016/j.foodres.2018.05.045
Dolara P, Luceri C, De Filippo C, Femia AP, Giovannelli L, Caderni G, Cecchini C, Silvi S, Orpianesi C, Cresci A (2005) Red wine polyphenols influence carcinogenesis, intestinal microflora, oxidative damage and gene expression profiles of colonic mucosa in F344 rats. Mutat Res 591(1–2):237–246. https://doi.org/10.1016/j.mrfmmm.2005.04.022
Viveros A, Chamorro S, Pizarro M, Arija I, Centeno C, Brenes A (2011) Effects of dietary polyphenol-rich grape products on intestinal microflora and gut morphology in broiler chicks. Poult Sci 90(3):566–578. https://doi.org/10.3382/ps.2010-00889
Molan A-L, Liu Z, Kruger M (2010) The ability of blackcurrant extracts to positively modulate key markers of gastrointestinal function in rats. World J Microb Biot 26(10):1735–1743. https://doi.org/10.1007/s11274-010-0352-4
Tzounis X, Rodriguez-Mateos A, Vulevic J, Gibson GR, Kwik-Uribe C, Spencer JP (2011) Prebiotic evaluation of cocoa-derived flavanols in healthy humans by using a randomized, controlled, double-blind, crossover intervention study. Am J Clin Nutr 93(1):62–72. https://doi.org/10.3945/ajcn.110.000075
Fritsch C, Heinrich V, Vogel RF, Toelstede S (2016) Phenolic acid degradation potential and growth behavior of lactic acid bacteria in sunflower substrates. Food Microb 57:178–186. https://doi.org/10.1016/j.fm.2016.03.003
Stead D (1994) The effect of chlorogenic, gallic and quinic acids on the growth of spoilage strains of Lactobacillus collinoides and Lactobacillus brevis. Lett Appl Microbiol 18(2):112–114. https://doi.org/10.1111/j.1472-765X.1994.tb00819.x
Parkar SG, Stevenson DE, Skinner MA (2008) The potential influence of fruit polyphenols on colonic microflora and human gut health. Int J Food Microbiol 124(3):295–298. https://doi.org/10.1016/j.ijfoodmicro.2008.03.017
Xia D, Wu X, Shi J, Yang Q, Zhang Y (2011) Phenolic compounds from the edible seeds extract of Chinese Mei (Prunus mume Sieb. et Zucc) and their antimicrobial activity. LWT Food Sci Technol 44(1):347–349. https://doi.org/10.1016/j.lwt.2010.05.017
Gotteland M, Andrews M, Toledo M, Munoz L, Caceres P, Anziani A, Wittig E, Speisky H, Salazar G (2008) Modulation of Helicobacter pylori colonization with cranberry juice and Lactobacillus johnsonii La1 in children. Nutrition 24(5):421–426. https://doi.org/10.1016/j.nut.2008.01.007
Vitali B, Ndagijimana M, Cruciani F, Carnevali P, Candela M, Guerzoni ME, Brigidi P (2010) Impact of a synbiotic food on the gut microbial ecology and metabolic profiles. BMC Microbiol 10:4. https://doi.org/10.1186/1471-2180-10-4
Piekarska-Radzik L, Klewicka E, Milala J, Klewicki R, Rosół N, Matysiak B, Sójka M, Markowski J (2019) Wpływ polifenoli z wytłoków z pseudoowoców Rosa rugosa Thunb. na wzrost bakterii z rodzaju Lactobacillus. Impact of polyphenols from Rosa rugosa Thunb. pseudofruits pomace on growth of Lactobacillus bacteria. ZNTJ 26:73–87. https://doi.org/10.15193/zntj/2019/120/298
China R, Mukherjee S, Sen S, Bose S, Datta S, Koley H, Ghosh S, Dhar P (2012) Antimicrobial activity of Sesbania grandiflora flower polyphenol extracts on some pathogenic bacteria and growth stimulatory effect on the probiotic organism Lactobacillus acidophilus. Microbiol Res 167(8):500–506. https://doi.org/10.1016/j.micres.2012.04.003
Cueva C, Sanchez-Patan F, Monagas M, Walton GE, Gibson GR, Martin-Alvarez PJ, Bartolome B, Moreno-Arribas MV (2013) In vitro fermentation of grape seed flavan-3-ol fractions by human faecal microbiota: changes in microbial groups and phenolic metabolites. FEMS Microbiol Ecol 83(3):792–805. https://doi.org/10.1111/1574-6941.12037
Das A, Datta S, Mukherjee S, Bose S, Ghosh S, Dhar P (2015) Evaluation of antioxidative, antibacterial and probiotic growth stimulatory activities of Sesamum indicum honey containing phenolic compounds and lignans. LWT Food Sci Technol 61(1):244–250. https://doi.org/10.1016/j.lwt.2014.11.044
Sarkar D, Ankolekar C, Pinto M, Shetty K (2015) Dietary functional benefits of Bartlett and Starkrimson pears for potential management of hyperglycemia, hypertension and ulcer bacteria Helicobacter pylori while supporting beneficial probiotic bacterial response. Food Res Int 69:80–90. https://doi.org/10.1016/j.foodres.2014.12.014
Coman M, Oancea A-M, Verdenelli C, Cecchini C, Bahrim GE, Orpianesi C, Cresci A, Silvi S (2018) Polyphenol content and in vitro evaluation of antioxidant, antimicrobial and prebiotic properties of red fruit extracts. Eur Food Res Technol 244(4):735–745. https://doi.org/10.1007/s00217-017-2997-9
Zhang X, Zhu X, Sun Y, Hu B, Sun Y, Jabbar S, Zeng X (2013) Fermentation in vitro of EGCG, GCG and EGCG3"Me isolated from Oolong tea by human intestinal microbiota. Food Res Int 54(2):1589–1595. https://doi.org/10.1016/j.foodres.2013.10.005
Santamaria L, Reveron I, Plaza-Vinuesa L, Oliveros JC, de Las RB, Munoz R, Lopez de Felipe F (2019) Oleuropein transcriptionally primes Lactobacillus plantarum to interact with plant hosts. Front Microbiol 10:2177. https://doi.org/10.3389/fmicb.2019.02177
Reveron I, Plaza-Vinuesa L, Franch M, de Las RB, Munoz R, Lopez de Felipe F (2018) Transcriptome-based analysis in Lactobacillus plantarum WCFS1 reveals new insights into resveratrol effects at system level. Mol Nutr Food Res 62(9):e1700992. https://doi.org/10.1002/mnfr.201700992
Funding
Not applicable.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Compliance with ethics requirements
This review does not contain any studies with human participants or animals performed by any of the authors.
Ethics approval
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Piekarska-Radzik, L., Klewicka, E. Mutual influence of polyphenols and Lactobacillus spp. bacteria in food: a review. Eur Food Res Technol 247, 9–24 (2021). https://doi.org/10.1007/s00217-020-03603-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00217-020-03603-y