Abstract
Autism spectrum disorder (ASD) is a broad and heterogeneous group of neurological developmental disorders characterized by impaired social interaction and communication, restricted and repetitive behavioural patterns, and altered sensory processing. Currently, no reliable ASD molecular biomarkers are available. Since immune dysregulation has been supposed to be related with ASD onset and dyslipidaemia has been recognized as an early symptom of biological perturbation, lipid extracts from peripheral blood mononuclear cells (PBMCs), consisting primarily of lymphocytes (T cells, B cells, and NK cells) and monocytes, of 38 children with ASD and their non-autistic siblings were investigated by hydrophilic interaction liquid chromatography (HILIC) coupled with electrospray ionization and Fourier-transform mass spectrometry (ESI-FTMS). Performances of two freeware software for data extraction and processing were compared with acquired reliable data regardless of the used informatics. A reduction of variables from 1460 by the untargeted XCMS to 324 by the semi-untargeted Alex123 software was attained. All-ion fragmentation (AIF) MS scans along with Alex123 software were successfully applied to obtain information related to fatty acyl chain composition of six glycerophospholipid classes occurring in PBMC. Principal component analysis (PCA) and partial least squares discriminant analysis (PLS-DA) were explored to verify the occurrence of significant differences in the lipid pool composition of ASD children compared with 36 healthy siblings. After rigorous statistical validation, we conclude that phospholipids extracted from PBMC of children affected by ASD do not exhibit diagnostic biomarkers. Yet interindividual variability comes forth from this study as the dominant effect in keeping with the existing phenotypic and etiological heterogeneity among ASD individuals.

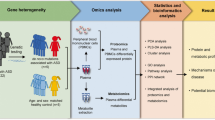
Graphical abstract







Similar content being viewed by others
References
American Psychiatric Association. Neurodevelopmental disorders. Diagn Stat Man Ment Disord. 2013. https://doi.org/10.1176/appi.books.9780890425596.dsm01.
Fombonne É. Epidemiological surveys of autism and other pervasive developmental disorders: an update. J Autism Dev Disord. 2003;33:365–82. https://doi.org/10.1023/A:1025054610557.
Daniels AM, Mandell DS. Explaining differences in age at autism spectrum disorder diagnosis: a critical review. Autism. 2014;18:583–97. https://doi.org/10.1177/1362361313480277.
Charman T (2015) Variability in neurodevelopmental disorders: evidence from autism spectrum disorders. In: Neurodev. Disord. Res. challenges Solut. Psychology Press, pp 117–140
Shen L, Liu XK, Zhang H, Lin J, Feng C, Iqbal J. Biomarkers in autism spectrum disorders: current progress. Clin Chim Acta. 2020;502:41–54. https://doi.org/10.1016/j.cca.2019.12.009.
Nguyen RL, Medvedeva YV, Ayyagari TE, Schmunk G, Gargus JJ. Intracellular calcium dysregulation in autism spectrum disorder: an analysis of converging organelle signaling pathways. Biochim Biophys Acta, Mol Cell Res. 2018;1865:1718–32. https://doi.org/10.1016/j.bbamcr.2018.08.003.
Palmieri L, Papaleo V, Porcelli V, Scarcia P, Gaita L, Sacco R, et al. Altered calcium homeostasis in autism-spectrum disorders: evidence from biochemical and genetic studies of the mitochondrial aspartate/glutamate carrier AGC1. Mol Psychiatry. 2010;15:38–52. https://doi.org/10.1038/mp.2008.63.
Palmieri L, Persico AM. Mitochondrial dysfunction in autism spectrum disorders: cause or effect? Biochim Biophys Acta Bioenerg. 2010;1797:1130–7. https://doi.org/10.1016/j.bbabio.2010.04.018.
Rose S, Niyazov DM, Rossignol DA, Goldenthal M, Kahler SG, Frye RE. Clinical and molecular characteristics of mitochondrial dysfunction in autism spectrum disorder. Mol Diagnosis Ther. 2018;22:571–93. https://doi.org/10.1007/s40291-018-0352-x.
Ashwood P, Wills S, Van de Water J. The immune response in autism: a new frontier for autism research. J Leukoc Biol. 2006;80:1–15. https://doi.org/10.1189/jlb.1205707.
Napolioni V, Persico AM, Porcelli V, Palmieri L. The mitochondrial aspartate/glutamate carrier AGC1 and calcium homeostasis: physiological links and abnormalities in autism. Mol Neurobiol. 2011;44:83–92. https://doi.org/10.1007/s12035-011-8192-2.
Sahin M, Sur M. Genes, circuits, and precision therapies for autism and related neurodevelopmental disorders. Science. 2015;350:aab3897–905. https://doi.org/10.1126/science.aab3897.
Gładysz D, Krzywdzińska A, Hozyasz KK. Immune abnormalities in autism spectrum disorder—could they hold promise for causative treatment? Mol Neurobiol. 2018;55:6387–435. https://doi.org/10.1007/s12035-017-0822-x.
Mead J, Ashwood P. Evidence supporting an altered immune response in ASD. Immunol Lett. 2015;163:49–55. https://doi.org/10.1016/j.imlet.2014.11.006.
Warren RP, Margaretten NC, Pace NC, Foster A. Immune abnormalities in patients with autism. J Autism Dev Disord. 1986;16:189–97. https://doi.org/10.1007/BF01531729.
Glatt SJ, Tsuang MT, Winn M, Chandler SD, Collins M, Lopez L, et al. Blood-based gene expression signatures of infants and toddlers with autism. J Am Acad Child Adolesc Psychiatry. 2012;51:934. https://doi.org/10.1016/j.jaac.2012.07.007.
Shen L, Feng C, Zhang K, Chen Y, Gao Y, Ke J, et al. Proteomics study of peripheral blood mononuclear cells (PBMCs) in autistic children. Front Cell Neurosci. 2019;13:105. https://doi.org/10.3389/fncel.2019.00105.
Losito I, Patruno R, Conte E, Cataldi TRI, Megli FM, Palmisano F. Phospholipidomics of human blood microparticles. Anal Chem. 2013;85:6405–13. https://doi.org/10.1021/ac400829r.
Cífková E, Holčapek M, Lísa M. Nontargeted lipidomic characterization of porcine organs using hydrophilic interaction liquid chromatography and off-line two-dimensional liquid chromatography-electrospray ionization mass spectrometry. Lipids. 2013;48:915–28. https://doi.org/10.1007/s11745-013-3820-4.
Ventura G, Bianco M, Calvano CD, Losito I, Cataldi TRI. HILIC-ESI-FTMS with all ion fragmentation (AIF) scans as a tool for fast lipidome investigations. Molecules. 2020;25:2310. https://doi.org/10.3390/molecules25102310.
Tsugawa H, Ikeda K, Arita M. The importance of bioinformatics for connecting data-driven lipidomics and biological insights. Biochim Biophys Acta Mol Cell Biol Lipids. 2017;1862:762–5. https://doi.org/10.1016/j.bbalip.2017.05.006.
Forsberg EM, Huan T, Rinehart D, Benton HP, Warth B, Hilmers B, et al. Data processing, multi-omic pathway mapping, and metabolite activity analysis using XCMS online. Nat Protoc. 2018;13:633–51. https://doi.org/10.1038/nprot.2017.151.
Husen P, Tarasov K, Katafiasz M, Sokol E, Vogt J, Baumgart J, et al. Analysis of lipid experiments (ALEX): a software framework for analysis of high-resolution shotgun lipidomics data. PLoS One. 2013;8:e79736. https://doi.org/10.1371/journal.pone.0079736.
Ellis SR, Paine MRL, Eijkel GB, Pauling JK, Husen P, Jervelund MW, et al. Automated, parallel mass spectrometry imaging and structural identification of lipids. Nat Methods. 2018;15:515–8. https://doi.org/10.1038/s41592-018-0010-6.
Liebisch G, Vizcaíno JA, Köfeler H, Trötzmüller M, Griffiths WJ, Schmitz G, et al. Shorthand notation for lipid structures derived from mass spectrometry. J Lipid Res. 2013;54:1523–30. https://doi.org/10.1194/jlr.M033506.
Liebisch G, Ekroos K, Hermansson M, Ejsing CS. Reporting of lipidomics data should be standardized. Biochim Biophys Acta Mol Cell Biol Lipids. 2017;1862:747–51. https://doi.org/10.1016/j.bbalip.2017.02.013.
Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–7. https://doi.org/10.1139/o59-099.
Dunn WB, Broadhurst D, Begley P, Zelena E, Francis-Mcintyre S, Anderson N, et al. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat Protoc. 2011;6:1060–83. https://doi.org/10.1038/nprot.2011.335.
Kamleh MA, Ebbels TMD, Spagou K, Masson P, Want EJ. Optimizing the use of quality control samples for signal drift correction in large-scale urine metabolic profiling studies. Anal Chem. 2012;84:2670–7. https://doi.org/10.1021/ac202733q.
Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B. 1995;57:289–300. https://doi.org/10.1111/j.2517-6161.1995.tb02031.x.
Granafei S, Azzone P, Spinelli VAVA, Losito I, Palmisano F, Cataldi TRITRI. Hydrophilic interaction and reversed phase mixed-mode liquid chromatography coupled to high resolution tandem mass spectrometry for polar lipids analysis. J Chromatogr A. 2016;1477:47–55. https://doi.org/10.1016/j.chroma.2016.11.048.
Granafei S, Losito I, Palmisano F, Cataldi TRI. Identification of isobaric lyso-phosphatidylcholines in lipid extracts of gilthead sea bream (Sparus aurata) fillets by hydrophilic interaction liquid chromatography coupled to high-resolution Fourier-transform mass spectrometry. Anal Bioanal Chem. 2015;407:6391–404. https://doi.org/10.1007/s00216-015-8671-9.
Calvano CD, Sardanelli AM, Ventura G, Glaciale M, Savino L, Losito I, et al. Development and use of advanced mass spectrometry techniques for the characterization of cellular and mitochondrial lipidomic profiling in control fibroblasts and Parkinson’s disease patients. Trends Pharm Biomed Anal. 2018;1:1–10. https://doi.org/10.15761/TPBA.1000102.
Calvano CD, Ventura G, Sardanelli AM, Savino L, Losito I, De Michele G, et al. Searching for potential lipid biomarkers of parkinson’s disease in parkin-mutant human skin fibroblasts by HILIC-ESI-MS/MS: preliminary findings. Int J Mol Sci. 2019;20:3341. https://doi.org/10.3390/ijms20133341.
Patti GJ, Tautenhahn R, Siuzdak G. Meta-analysis of untargeted metabolomic data from multiple profiling experiments. Nat Protoc. 2012;7:508–16. https://doi.org/10.1038/nprot.2011.454.
Libiseller G, Dvorzak M, Kleb U, Gander E, Eisenberg T, Madeo F, et al. IPO: a tool for automated optimization of XCMS parameters. BMC Bioinf. 2015;16:1–10. https://doi.org/10.1186/s12859-015-0562-8.
Albóniga OE, González O, Alonso RM, Xu Y, Goodacre R. Optimization of XCMS parameters for LC–MS metabolomics: an assessment of automated versus manual tuning and its effect on the final results. Metabolomics. 2020;16:14. https://doi.org/10.1007/s11306-020-1636-9.
Kuhl C, Tautenhahn R, Böttcher C, Larson TR, Neumann S. CAMERA: an integrated strategy for compound spectra extraction and annotation of liquid chromatography/mass spectrometry data sets. Anal Chem. 2012;84:283–9. https://doi.org/10.1021/ac202450g.
Han X (2016) Factors affecting accurate quantification of lipids. In: Wiley (ed) Lipidomics Compr. Mass Spectrom. Lipids. John Wiley & Sons, Inc, Hoboken, pp 335–352.
Dunn WB, Wilson ID, Nicholls AW, Broadhurst D. The importance of experimental design and QC samples in large-scale and MS-driven untargeted metabolomic studies of humans. Bioanalysis. 2012;4:2249–64. https://doi.org/10.4155/bio.12.204.
Broadhurst D, Goodacre R, Reinke SN, Kuligowski J, Wilson ID, Lewis MR, et al. Guidelines and considerations for the use of system suitability and quality control samples in mass spectrometry assays applied in untargeted clinical metabolomic studies. Metabolomics. 2018;14:0. https://doi.org/10.1007/s11306-018-1367-3.
Bartel J, Krumsiek J, Theis FJ. Statistical methods for the analysis of high-throughput metabolomics data. Comput Struct Biotechnol J. 2013;4:e201301009. https://doi.org/10.5936/csbj.201301009.
Brereton RG (2003) Chemometrics. doi: https://doi.org/10.1002/0470863242.
Bohm HV, Stewart MG. Brief report: on the concordance percentages for autistic spectrum disorder of twins. J Autism Dev Disord. 2009;39:806–8. https://doi.org/10.1007/s10803-008-0683-2.
Gromski PS, Muhamadali H, Ellis DI, Xu Y, Correa E, Turner ML, et al. A tutorial review: metabolomics and partial least squares-discriminant analysis - a marriage of convenience or a shotgun wedding. Anal Chim Acta. 2015;879:10–23. https://doi.org/10.1016/j.aca.2015.02.012.
Chauhan V, Chauhan A, Cohen IL, Brown WT, Sheikh A. Alteration in amino-glycerophospholipids levels in the plasma of children with autism: a potential biochemical diagnostic marker. Life Sci. 2004;74:1635–43. https://doi.org/10.1016/j.lfs.2003.08.024.
Murphy R (2015) Tandem mass spectrometry of lipids
Sumner LW, Amberg A, Barrett D, Beale MH, Beger R, Daykin CA, et al. Proposed minimum reporting standards for chemical analysis. Metabolomics. 2007;3:211–21. https://doi.org/10.1007/s11306-007-0082-2.
Storey JD. A direct approach to false discovery rates. J R Stat Soc Ser B (Stat Methodol). 2002;64:479–98. https://doi.org/10.1111/1467-9868.00346.
Wenk MR. The emerging field of lipidomics. Nat Rev Drug Discov. 2005;4:594–610. https://doi.org/10.1038/nrd1776.
Harayama T, Riezman H. Understanding the diversity of membrane lipid composition. Nat Rev Mol Cell Biol. 2018;19:281–96. https://doi.org/10.1038/nrm.2017.138.
Han X. Lipids and lipidomics. In: Lipidomics. Hoboken: John Wiley & Sons, Inc; 2016. p. 1–20.
Calvano CD, Palmisano F, Cataldi TRI. Understanding neurodegenerative disorders by MS-based lipidomics. Bioanalysis. 2018;10:787–90. https://doi.org/10.4155/bio-2018-0023.
Yang K, Han X. Accurate quantification of lipid species by electrospray ionization mass spectrometry - meets a key challenge in lipidomics. Metabolites. 2011;1:21–40. https://doi.org/10.3390/metabo1010021.
Wiest MM, German JB, Harvey DJ, Watkins SM, Hertz-Picciotto I. Plasma fatty acid profiles in autism: a case-control study. Prostaglandins Leukot Essent Fat Acids. 2009;80:221–7. https://doi.org/10.1016/j.plefa.2009.01.007.
Brigandi SA, Shao H, Qian SY, Shen Y, Wu BL, Kang JX. Autistic children exhibit decreased levels of essential fatty acids in red blood cells. Int J Mol Sci. 2015;16:10061–76. https://doi.org/10.3390/ijms160510061.
Richardson AJ, Ross MA. Fatty acid metabolism in neurodevelopmental disorder: a new perspective on associations between attention-deficit/hyperactivity disorder, dyslexia, dyspraxia and the autistic spectrum. Prostaglandins Leukot Essent Fat Acids. 2000;63:1–9. https://doi.org/10.1054/plef.2000.0184.
El-Ansary A, Chirumbolo S, Bhat RS, Dadar M, Ibrahim EM, Bjørklund G. The role of lipidomics in autism spectrum disorder. Mol Diagnosis Ther. 2020;24:31–48. https://doi.org/10.1007/s40291-019-00430-0.
Vancassel S, Durand G, Barthélémy C, Lejeune B, Martineau J, Guilloteau D, et al. Plasma fatty acid levels in autistic children. Prostaglandins Leukot Essent Fat Acids. 2001;65:1–7. https://doi.org/10.1054/plef.2001.0281.
Bell JG, MacKinlay EE, Dick JR, MacDonald DJ, Boyle RM, Glen ACA (2004) Essential fatty acids and phospholipase A2 in autistic spectrum disorders. In: Prostaglandins Leukot. Essent. Fat. Acids. pp 201–204
Bell JG, Miller D, MacDonald DJ, MacKinlay EE, Dick JR, Cheseldine S, et al. The fatty acid compositions of erythrocyte and plasma polar lipids in children with autism, developmental delay or typically developing controls and the effect of fish oil intake. Br J Nutr. 2010;103:1160–7. https://doi.org/10.1017/S0007114509992881.
Bu B, Ashwood P, Harvey D, King IB, Van de Water J, Jin LW. Fatty acid compositions of red blood cell phospholipids in children with autism. Prostaglandins Leukot Essent Fat Acids. 2006;74:215–21. https://doi.org/10.1016/j.plefa.2006.02.001.
El-Ansary AK, Ben Bacha AG, Al-Ayahdi LY. Plasma fatty acids as diagnostic markers in autistic patients from Saudi Arabia. Lipids Health Dis. 2011;10:62. https://doi.org/10.1186/1476-511X-10-62.
Howsmon DP, Adams JB, Kruger U, Geis E, Gehn E, Hahn J. Erythrocyte fatty acid profiles in children are not predictive of autism spectrum disorder status: a case control study. Biomark Res. 2018;6:12. https://doi.org/10.1186/s40364-018-0125-z.
Neubronner J, Schuchardt JP, Kressel G, Merkel M, Von Schacky C, Hahn A. Enhanced increase of omega-3 index in response to long-term n-3 fatty acid supplementation from triacylglycerides versus ethyl esters. Eur J Clin Nutr. 2011;65:247–54. https://doi.org/10.1038/ejcn.2010.239.
Al-Farsi YM, Waly MI, Deth RC, Al-Sharbati MM, Al-Shafaee M, Al-Farsi O, et al. Impact of nutrition on serum levels of docosahexaenoic acid among Omani children with autism. Nutrition. 2013;29:1142–6. https://doi.org/10.1016/j.nut.2013.03.009.
De Rubeis S, Buxbaum JD. Genetics and genomics of autism spectrum disorder: embracing complexity. Hum Mol Genet. 2015;24:R24–31. https://doi.org/10.1093/hmg/ddv273.
Sandin S, Lichtenstein P, Kuja-Halkola R, Larsson H, Hultman CM, Reichenberg A. The familial risk of autism. JAMA. 2014;311:1770. https://doi.org/10.1001/jama.2014.4144.
Ng M, de Montigny JG, Ofner M, Do M. Environmental factors associated with autism spectrum disorder: a scoping review for the years 2003–2013. Health Promot Chronic Dis Prev Can. 2017;37:1–23. https://doi.org/10.24095/hpcdp.37.1.01.
Calder PC. The relationship between the fatty acid composition of immune cells and their function. Prostaglandins Leukot Essent Fat Acids. 2008;79:101–8. https://doi.org/10.1016/j.plefa.2008.09.016.
Funding
This work was supported by the following projects: (i) PONa3_00395/1 ‘BIOSCIENZE & SALUTE (B&H)’ and (ii) SIR 2014, Identification and Characterization of Biomarkers for Autism Spectrum Disorder of ‘Ministero per l’Istruzione, l’Università e la Ricerca’ (MIUR). G.V. during his Ph.D. period abroad carried out part of this work in Manchester at the Institute of Biotechnology.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(PDF 1027 kb)
Rights and permissions
About this article
Cite this article
Ventura, G., Calvano, C.D., Porcelli, V. et al. Phospholipidomics of peripheral blood mononuclear cells (PBMCs): the tricky case of children with autism spectrum disorder (ASD) and their healthy siblings. Anal Bioanal Chem 412, 6859–6874 (2020). https://doi.org/10.1007/s00216-020-02817-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-020-02817-z