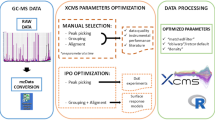
Abstract
Introduction
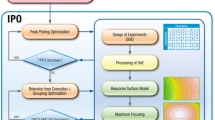
Several software packages containing diverse algorithms are available for processing Liquid Chromatography-Mass Spectrometry (LC–MS) chromatographic data and within these deconvolution packages different parameters settings can lead to different outcomes. XCMS is the most widely used peak picking and deconvolution software for metabolomics, but the parameter selection can be hard for inexpert users. To solve this issue, the automatic optimization tools such as Isotopologue Parameters Optimization (IPO) can be extremely helpful.
Objectives
To evaluate the suitability of IPO as a tool for XCMS parameters optimization and compare the results with those manually obtained by an exhaustive examination of the LC–MS characteristics and performance.
Methods
Raw HPLC-TOF–MS data from two types of biological samples (liver and plasma) analysed in both positive and negative electrospray ionization modes from three groups of piglets were processed with XCMS using parameters optimized following two different approaches: IPO and Manual. The outcomes were compared to determine the advantages and disadvantages of using each method.
Results
IPO processing produced the higher number of repeatable (%RSD < 20) and significant features for all data sets and allowed the different piglet groups to be distinguished. Nevertheless, on multivariate level, similar clustering results were obtained by Principal Component Analysis (PCA) when applied to IPO and manual matrices.
Conclusion
IPO is a useful optimization tool that helps in choosing the appropriate parameters. It works well on data with a good LC–MS performance but the lack of such adequate data can result in unrealistic parameter settings, which might require further investigation and manual tuning. On the contrary, manual selection criteria requires deeper knowledge on LC–MS, programming language and XCMS parameter interpretation, but allows a better fine-tuning of the parameters, and thus more robust deconvolution.



Similar content being viewed by others
Data availability
The datasets generated and analysed during the current study are available from the corresponding author on request.
References
Broadhurst, D., Goodacre, R., Reinke, S. N., Kuligowski, J., Wilson, I. D., Lewis, M. R., et al. (2018). Guidelines and considerations for the use of system suitability and quality control samples in mass spectrometry assays applied in untargeted clinical metabolomic studies. Metabolomics, 14(6), 72.
Dryden, I. L., & Mardia, K. V. (1998). Statistical shape analysis. Wiley series in probability and statistics. Chichester: Wiley.
Dudzik, D., Barbas-Bernardos, C., Garcia, A., & Barbas, C. (2018). Quality assurance procedures for mass spectrometry untargeted metabolomics. A review. Journal of Pharmaceutical and Biomedical Analysis, 147, 149–173.
Dunn, W. B., Broadhurst, D., Begley, P., Zelena, E., Francis-McIntyre, S., Anderson, N., et al. (2011). Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nature Protocols, 6(7), 1060–1083.
Grimbs, A., Shrestha, A., Rezk Ahmed, S. D., Grimbs, S., Hakeem, S. I., Hutt, M., et al. (2017). Bioactivity in rhododendron: A systemic analysis of antimicrobial and cytotoxic activities and their phylogenetic and phytochemical origins. Frontiers in Plant Science, 8, 551.
Harvey Colin, J. B., Schlecht, U., Horecka, J., Fischer, C. R., Li, J., Naughton, B., et al. (2018). HEx: A heterologous expression platform for the discovery of fungal natural products. Science Advances, 4(4),eaar5459.
Kirwan, J., Broadhurst, D., Davidson, R., & Viant, M. (2013). Characterising and correcting batch variation in an automated direct infusion mass spectrometry (DIMS) metabolomics workflow. Analytical and Bioanalytical Chemistry, 405(15), 5147–5157.
Kuhl, C., Tautenhahn, R., Boettcher, C., Larson, T. R., & Neumann, S. (2012). CAMERA: An integrated strategy for compound spectra extraction and annotation of liquid chromatography/mass spectrometry data sets. Analytical Chemistry (Washington, DC), 84(1), 283–289.
Lazar, G., Florina, R., Socaciu, M., & Socaciu, C. (2015). Bioinformatics tools for metabolomic data processing and analysis using untargeted liquid chromatography coupled with mass spectrometry. Bulletin of University of Agricultural Sciences and Veterinary Medicine Cluj-Napoca Animal Science and Biotechnologies. https://doi.org/10.15835/buasvmcn-asb:11536.
Libiseller, G., Dvorzak, M., Kleb, U., Gander, E., Eisenberg, T., Madeo, F., et al. (2015). IPO: A tool for automated optimization of XCMS parameters. BMC Bioinformatics, 16, 1–10.
Myers, O. D., Sumner, S. J., Li, S., Barnes, S., & Du, X. (2017). Detailed investigation and comparison of the XCMS and MZmine 2 chromatogram construction and chromatographic peak detection methods for preprocessing mass spectrometry metabolomics data. Analytical Chemistry (Washington, DC), 89(17), 8689–8695.
Narath, S. H., Mautner, S. I., Svehlikova, E., Schultes, B., Pieber, T. R., Sinner, F. M., et al. (2016). An untargeted metabolomics approach to characterize short-term and long-term metabolic changes after bariatric surgery. PLoS ONE, 11(9), e0161425/1–e0161425/18.
Prince, J. T., & Marcotte, E. M. (2006). Chromatographic alignment of ESI-LC-MS proteomics data sets by ordered bijective interpolated warping. Analytical Chemistry, 78(17), 6140–6152.
Roszkowska, A., Yu, M., Bessonneau, V., Bragg, L., Servos, M., & Pawliszyn, J. (2018). Tissue storage affects lipidome profiling in comparison to in vivo microsampling approach. Science Reports, 8(1), 1–10.
Smith, C. A., Want, E. J., O'Maille, G., Abagyan, R., & Siuzdak, G. (2006). XCMS: Processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Analytical Chemistry, 78(3), 779–787.
Spicer, R., Salek, R. M., Moreno, P., Canueto, D., & Steinbeck, C. (2017). Navigating freely-available software tools for metabolomics analysis. Metabolomics, 13(9), 1–16.
Stoessel, D., Schauer, N., Stoessel, D., Walther, D., Stoessel, D., Walther, D., et al. (2018a). Promising metabolite profiles in the plasma and CSF of early clinical Parkinson's disease. Frontiers in Aging Neuroscience, 10, 51.
Stoessel, D., Schauer, N., Stoessel, D., Walther, D., Stoessel, D., Walther, D., et al. (2018b). Metabolomic profiles for primary progressive multiple sclerosis stratification and disease course monitoring. Frontiers in Human Neuroscience, 12, 226.
Tautenhahn, R., Boettcher, C., & Neumann, S. (2008). Highly sensitive feature detection for high resolution LC/MS. BMC Bioinformatics, 9, 504.
Tautenhahn, R., Patti, G. J., Rinehart, D., & Siuzdak, G. (2012). XCMS online: A web-based platform to process untargeted metabolomic data. Analytical Chemistry (Washington, DC), 84(11), 5035–5039.
Weber, R. J. M., Lawson, T. N., Salek, R. M., Ebbels, T. M. D., Glen, R. C., Goodacre, R., et al. (2017). Computational tools and workflows in metabolomics: An international survey highlights the opportunity for harmonisation through galaxy. Metabolomics, 13(2), 1–5.
Acknowledgements
Authors thank the Experimental Neonatal Physiology Unit of the BioCruces Health Research Institute (Cruces University Hospital, Basque Country, Spain) for collecting and providing the samples. O.E.A. thanks the Ministry of Economy and Competitiveness for her predoctoral contract. Authors thank for technical and human support provided by SGIker of UPV/EHU and European funding (ERDF and ESF).
Funding
This research was funded by UPV/EHU (Project GIU16/04), the Spanish Ministry of Economy and Competitiveness (Project CTQ2013-46179-R) and the UK Wellcome Trust for MetaboFlow (Grant 202952/Z/16/Z).
Author information
Authors and Affiliations
Contributions
The following authors contributed to conception and design of the study: O.E.A., O.G., R.M.A., Y.X. and R.G.; funding acquisition: R.M.A; experimental performance and acquisition of data: O.E.A. and O.G.; data analysis: O.E.A. and Y.X.; writing – original draft preparation; O.E.A. and O.G., and supervision: O.G., R.M.A., Y.X. and R.G. All authors revised the article critically for important intellectual content and approved the final version.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interest.
Ethical approval
The study was approved by the Ethical Committee for Animal Welfare following the European and Spanish regulations for protection of experimental animals (86/609/EFC and RD 1201/2005).
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Albóniga, O.E., González, O., Alonso, R.M. et al. Optimization of XCMS parameters for LC–MS metabolomics: an assessment of automated versus manual tuning and its effect on the final results. Metabolomics 16, 14 (2020). https://doi.org/10.1007/s11306-020-1636-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11306-020-1636-9