Abstract
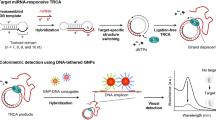
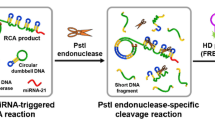
In this study, a biosensing system based on nicking-enhanced rolling circle amplification (N-RCA) was proposed for the highly sensitive detection of cancer-related let-7a microRNA (miRNA). The sensing system consists of a padlock probe (PP), which contains a target recognition sequence and two binding sites for nicking endonuclease (NEase), and molecular beacon (MB) as reporting molecule. Upon hybridization with let-7a, the PP can be circularized by ligase. Then, the miRNA acted as polymerization primer to initiate rolling circle amplification (RCA). With the assistance of NEase, RCA products can be nicked on the cyclized PP and are displaced during the subsequent duplication process, generating numerous nicked fragments (NFs). These NFs not only induce another RCA reaction but also open the molecular beacons (MBs) via hybridization, leading to significantly amplified fluorescence signal. Under the optimized conditions, this method exhibits high sensitivity toward target miRNA let-7a with a detection limit of as low as 10 pM, a dynamic range of three orders of magnitude is achieved, and its family member is easily distinguished even with only one mismatched base. Meanwhile, it displays good recovery and satisfactory reproducibility in fetal bovine serum (FBS). Therefore, these merits endow the newly proposed N-RCA strategy with powerful implications for miRNA detection.

A biosensing system based on nicking-enhanced rolling circle amplification (N-RCA) for the highly sensitive detection of cancer-related let-7a microRNA





Similar content being viewed by others
References
Friedman RC, Farh KK-H, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105.
Liang Y, Ridzon D, Wong L, Chen C. Characterization of microRNA expression profiles in normal human tissues. BMC Genomics. 2007;8:166.
Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–47.
Van Kouwenhove M, Kedde M, Agami R. MicroRNA regulation by RNA-binding proteins and its implications for cancer. Nat Rev Cancer. 2011;11:644–56.
Belgardt B-F, Ahmed K, Spranger M, Latreille M, Denzler R, Kondratiuk N, et al. The microRNA-200 family regulates pancreatic beta cell survival in type 2 diabetes. Nat Med. 2015;21:619–27.
Seronde M-F, Vausort M, Gayat E, Goretti E, Ng LL, Squire IB, et al. Circulating microRNAs and outcome in patients with acute heart failure. PLoS One. 2015;10:e0142237.
Jeanson-Leh L, Lameth J, Krimi S, Buisset J, Amor F, Le Guiner C, et al. Serum profiling identifies novel muscle miRNA and cardiomyopathy-related miRNA biomarkers in Golden Retriever muscular dystrophy dogs and Duchenne muscular dystrophy patients. Am J Pathol. 2014;184:2885–98.
Li J, Tan S, Kooger R, Zhang C, Zhang Y. MicroRNAs as novel biological targets for detection and regulation. Chem Soc Rev. 2014;43:506–17.
Brueckner B, Stresemann C, Kuner R, Mund C, Musch T, Meister M, et al. The human let-7a-3 locus contains an epigenetically regulated microRNA gene with oncogenic function. Cancer Res. 2007;67:1419–23.
Válóczi A, Hornyik C, Varga N, Burgyán J, Kauppinen S, Havelda Z. Sensitive and specific detection of microRNAs by northern blot analysis using LNA-modified oligonucleotide probes. Nucleic Acids Res. 2004;32:e175.
Cissell KA, Shrestha S, Deo SK. MicroRNA detection: challenges for the analytical chemist. Anal Chem. 2007;79:4754–61.
Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, et al. Real-time quantification of microRNAs by stem–loop RT–PCR. Nucleic Acids Res. 2005;33:e179.
Benes V, Castoldi M. Expression profiling of microRNA using real-time quantitative PCR, how to use it and what is available. Methods. 2010;50:244–9.
Zhang J, Li Z, Wang H, Wang Y, Jia H, Yan J. Ultrasensitive quantification of mature microRNAs by real-time PCR based on ligation of a ribonucleotide-modified DNA probe. Chem Commun. 2011;47:9465–7.
Thomson JM, Parker J, Perou CM, Hammond SM. A custom microarray platform for analysis of microRNA gene expression. Nat Methods. 2004;1:47–53.
Lee JM, Jung Y. Two-temperature hybridization for microarray detection of label-free microRNAs with attomole detection and superior specificity. Angew Chem Int Edit. 2011;50:12487–90.
Harcourt EM, Kool ET. Amplified microRNA detection by templated chemistry. Nucleic Acids Res. 2012;40:e65.
Baker MB, Bao G, Searles CD. In vitro quantification of specific microRNA using molecular beacons. Nucleic Acids Res. 2011;40:e13.
Huang J, Su X, Li Z. Enzyme-free and amplified fluorescence DNA detection using bimolecular beacons. Anal Chem. 2012;84:5939–43.
Xu H, Zhang R, Li F, Zhou Y, Peng T, Wang X, et al. Double-hairpin molecular-beacon-based amplification detection for gene diagnosis linked to cancer. Anal Bioanal Chem. 2016;408:6181–8.
Xu J, Wu ZS, Wang Z, Le J, Zheng T, Jia L. Autonomous assembly of ordered metastable DNA nanoarchitecture and in situ visualizing of intracellular microRNAs. Biomaterials. 2017;120:57–65.
Xu J, Li H, Wu ZS, Qian J, Xue C, Jia L. Double-stem hairpin probe and ultrasensitive colorimetric detection of cancer-related nucleic acids. Theranostics. 2016;6:318–27.
Xu H, Xue C, Zhang R, Chen Y, Li F, Shen Z, et al. Exponential rolling circle amplification and its sensing application for highly sensitive DNA detection of tumor suppressor gene. Sensors Actuators B Chem. 2017;243:1240–7.
Li F, Zhao H, Wang Z-Y, Wu Z-S, Yang Z, Li C-C, et al. Single palindromic molecular beacon-based amplification for genetic analysis of cancers. Biosens Bioelectron. 2017;91:692–8.
Shen Z-F, Li F, Jiang Y-F, Chen C, Xu H, Li C-C, et al. Palindromic molecule beacon-based cascade amplification for colorimetric detection of cancer genes. Anal Chem. 2018;90:3335–40.
Zhou H, Xie SJ, Zhang SB, Shen GL, Yu RQ, Wu ZS. Isothermal amplification system based on template-dependent extension. Chem Commun. 2013;49:2448–50.
Bi S, Cui Y, Dong Y, Zhang N. Target-induced self-assembly of DNA nanomachine on magnetic particle for multi-amplified biosensing of nucleic acid, protein, and cancer cell. Biosens Bioelectron. 2014;53:207–13.
Xu H, Wu D, Zhang Y, Shi H, Ouyang C, Li F, et al. RCA-enhanced multifunctional molecule beacon-based strand-displacement amplification for sensitive microRNA detection. Sensors Actuators B Chem. 2018;258:470–7.
Wang Z-Y, Li F, Zhang Y, Zhao H, Xu H, Wu Z-S, et al. Sensitive detection of cancer gene based on a nicking-mediated RCA of circular DNA nanomachine. Sensors Actuators B Chem. 2017;251:692–8.
Ge J, Dong Z-Z, Bai D-M, Zhang L, Hu Y-L, Ji D-Y, et al. A novel label-free fluorescent molecular beacon for the detection of 3′–5′ exonuclease enzymatic activity using DNA-templated copper nanoclusters. New J Chem. 2017;41:9718–23.
Huang Z-M, Cai Q-Y, Ding D-C, Ge J, Hu Y-L, Yang J, et al. A facile label-free colorimetric method for highly sensitive glutathione detection by using manganese dioxide nanosheets. Sensors Actuators B Chem. 2017;242:355–61.
Luo C, Tang H, Cheng W, Yan L, Zhang D, Ju H, et al. A sensitive electrochemical DNA biosensor for specific detection of Enterobacteriaceae bacteria by exonuclease III-assisted signal amplification. Biosens Bioelectron. 2013;48:132–7.
Wang M, Fu Z, Li B, Zhou Y, Yin H, Ai S. One-step, ultrasensitive, and electrochemical assay of microRNAs based on T7 exonuclease assisted cyclic enzymatic amplification. Anal Chem. 2014;86:5606–10.
Wu Z-S, Zhou H, Zhang S, Shen G, Yu R. Electrochemical aptameric recognition system for a sensitive protein assay based on specific target binding-induced rolling circle amplification. Anal Chem. 2010;82:2282–9.
Chapin SC, Doyle PS. Ultrasensitive multiplexed microRNA quantification on encoded gel microparticles using rolling circle amplification. Anal Chem. 2011;83:7179–85.
Nilsson M, Antson DO, Barbany G, Landegren U. RNA-templated DNA ligation for transcript analysis. Nucleic Acids Res. 2001;29:578–81.
Qiu L, Qiu L, Zhou H, Wu Z, Shen G, Yu R. Sensitive and selective electrochemical DNA sensor for the analysis of cancer-related single nucleotide polymorphism. New J Chem. 2014;38:4711–5.
Wu ZS, Jiang JH, Shen GL, Yu RQ. Highly sensitive DNA detection and point mutation identification: an electrochemical approach based on the combined use of ligase and reverse molecular beacon. Hum Mutat. 2010;28:630–7.
Yu J, Li B, Milligan JN, Bhadra S, Ellington AD. Real-time detection of isothermal amplification reactions with thermostable catalytic hairpin assembly. J Am Chem Soc. 2013;135:7430–3.
Liu H, Li L, Duan L, Wang X, Xie Y, Tong L, et al. High specific and ultrasensitive isothermal detection of microRNA by padlock probe-based exponential rolling circle amplification. Anal Chem. 2013;85:7941–7.
Feng K, Qiu LP, Yang Y, Wu ZS, Shen GL, Yu RQ. Label-free optical bifunctional oligonucleotide probe for homogeneous amplification detection of disease markers. Biosens Bioelectron. 2011;29:66–75.
Dong H, Ma J, Wang J, Wu ZS, Sinko PJ, Jia L. A biofunctional molecular beacon for detecting single base mutations in cancer cells. Mol Ther Nucl Acids. 2016;5:e302.
Zhang S, Wu Z, Shen G, Yu R. A label-free strategy for SNP detection with high fidelity and sensitivity based on ligation-rolling circle amplification and intercalating of methylene blue. Biosens Bioelectron. 2009;24:3201–7.
Deng R, Tang L, Tian Q, Wang Y, Lin L, Li J. Toehold-initiated rolling circle amplification for visualizing individual microRNAs in situ in single cells. Angew Chem Int Edit. 2014;53:2389–93.
Simms D, Cizdziel PE, Chomczynski P. TRIzol: a new reagent for optimal single-step isolation of RNA. Focus. 1993;15:532–5.
Hu J, Wen C-Y, Zhang Z-L, Xie M, Hu J, Wu M, et al. Optically encoded multifunctional nanospheres for one-pot separation and detection of multiplex DNA sequences. Anal Chem. 2013;85:11929–35.
Cheng Y, Zhang X, Li Z, Jiao X, Wang Y, Zhang Y. Highly sensitive determination of microRNA using target-primed and branched rolling-circle amplification. Angew Chem Int Edit. 2009;121:3318–22.
Hikichi M, Kidokoro M, Haraguchi T, Iba H, Shida H, Tahara H, et al. MicroRNA regulation of glycoprotein B5R in oncolytic vaccinia virus reduces viral pathogenicity without impairing its antitumor efficacy. Mol Ther. 2011;19:1107–15.
Chen, Lijia, Xing, Yuan, Kehui, Wang, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006.
Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–14.
Funding
This work was supported by the National Natural Science Foundation of China (NSFC) (grant no: 21775024) and Zhejiang Province Natural Science Foundation of China (LY16C07002).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
This study was approved by the ethical review committee of The First Affiliated Hospital of Wenzhou. Written informed consent was obtained by the volunteer the human serum sample was obtained from.
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
ESM 1
(PDF 190 kb)
Rights and permissions
About this article
Cite this article
Gao, Z., Wu, C., Lv, S. et al. Nicking-enhanced rolling circle amplification for sensitive fluorescent detection of cancer-related microRNAs. Anal Bioanal Chem 410, 6819–6826 (2018). https://doi.org/10.1007/s00216-018-1277-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-018-1277-2