Abstract
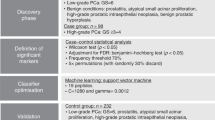
Cancer is responsible for millions of deaths worldwide, but most base diseases may be cured if detected early. Screening tests may be used to identify early-stage malignant neoplasms. However, the major screening tool for prostate cancer, the prostate-specific antigen test, has unsuitable sensitivity. Since cancer cells may affect the pattern of consumption and excretion of nucleosides, such biomolecules are putative biomarkers that can be used for diagnosis and treatment evaluation. Using a previously validated method for the analysis of nucleosides in blood serum by capillary electrophoresis with UV–vis spectroscopy detection, we investigated 60 samples from healthy individuals and 42 samples from prostate cancer patients. The concentrations of nucleosides in both groups were compared and a multivariate partial least squares–discriminant analysis classification model was optimized for prediction of prostate cancer. The validation of the model with an independent sample set resulted in the correct classification of 82.4% of the samples, with sensitivity of 90.5% and specificity of 76.7%. A significant downregulation of 5-methyluridine and inosine was observed, which can be indicative of the carcinogenic process. Therefore, such analytes are potential candidates for prostate cancer screening.

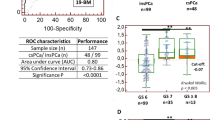
Separation of the studied nucleosides and the internal standard 8-Bromoguanosine by CE-UV (a); classification of the external validation samples (30 from healthy volunteers and 21 from prostate cancer patients) by the developed Partial Least Square – Discriminant Analysis (PLS-DA) model with accuracy of 82.4% (b); Receiver Operating Characteristics (ROC) curve (c); and Variable Importance in the Projection (VIP) values for the studied nucleosides (d). A significant down-regulation of 5- methyluridine (5mU) and inosine (I) was observed, which can be indicative of the presence of prostate tumors.







Similar content being viewed by others
References
World Health Organization (2014). Word Health Organization. http://www.who.int. Accessed 18 Mar 2014
Ruddon RW. Cancer Biology. 4th ed. New York: Oxford University Press; 2007.
Greave M. Cancer: the evolutionary legacy. 1st ed. New York: Oxford University Press; 2000.
Naylor S. Biomarkers: current perspectives and future prospects. Expert Rev Mol Diagn. 2003;3:525–9.
Atkinson AJ, Wayne AC, DeGruttola VG, et al. Biomarkers and surrogate endpoints: Preferred definitions and conceptual framework. Clin Pharmacol Ther. 2001;69:89–95.
Ramautar R, Somsen GW, de Jong GJ. CE-MS for metabolomics: developments and applications in the period 2010-2012. Electrophoresis. 2013;34:86–98.
Jain KK. The handbook of biomarkers. New York: Humana; 2010.
Vaidya VS, Bonventre JV, editors. Biomarkers in medicine, drug discovery and enviromental health, 1st ed. Hoboken: Wiley; 2010.
Bloomfield VA. Nucleic acids: stuctures, properties and functions. Sausalito: University Science Books; 2000.
Schram KH. Urinary nucleosides. Mass Spectrom Rev. 1998;17:131–251.
Markuszewski MJ, Struck W, Waszczuk-Jankowska M, Kaliszan R. Metabolomic approach for determination of urinary nucleosides as potential tumor markers using electromigration techniques. Electrophoresis. 2010;31:2300–10.
Bullinger D, Fröhlich H, Klaus F, et al. Bioinformatical evaluation of modified nucleosides as biomedical markers in diagnosis of breast cancer. Anal Chim Acta. 2008;618:29–34.
Cho SH, Jung BH, Lee SH, Lee WY, Kong G, Chung BC. Direct determination of nucleosides in the urine of patients with breast cancer using column-switching liquid chromatography-tandem mass spectrometry. Biomed Chromatogr. 2006;20:1229–36.
Djukovic D, Baniasadi HR, Kc R, Hammoud Z, Raftery D. Targeted serum metabolite profiling of nucleosides in esophageal adenocarcinoma. Rapid Commun Mass Spectrom. 2010;24:3057–62.
Feng B, Zheng MH, Zheng YF, Lu AG, Li JW, Wang ML, et al. Normal and modified urinary nucleosides represent novel biomarkers for colorectal cancer diagnosis and surgery monitoring. J Gastroenterol Hepatol. 2005;20:1913–9.
Fischbein A, Sharma OK, Selikoff IJ, Borek E. Urinary excretion of modified nucleosides in patients with malignant mesothelioma. Cancer Res. 1983;43:2971–4.
García-Closas M, Malats N, Real FX, et al. Genetic variation in the nucleotide excision repair pathway and bladder cancer risk. Cancer Epidemiol Biomarkers Prev. 2006;15:536–42.
Henneges C, Bullinger D, Fux R, et al. Prediction of breast cancer by profiling of urinary RNA metabolites using support vector machine-based feature selection. BMC Cancer. 2009;9:104–14.
Hsu WY, Chen CJ, Huang YC, Tsai FJ, Bin JL, Lai CC. Urinary nucleosides as biomarkers of breast, colon, lung, and gastric cancer in Taiwanese. PLoS One. 2013;8, e81701.
Jeng LB, Lo WY, Hsu WY, Lin WD, Lin CT, Lai CC, et al. Analysis of urinary nucleosides as helper tumor markers in hepatocellular carcinoma diagnosis. Rapid Commun Mass Spectrom. 2009;23:1543–9.
Kim KR, La S, Kim A, Kim JH, Liebich HM. Capillary electrophoretic profiling and pattern recognition analysis of urinary nucleosides from uterine myoma and cervical cancer patients. J Chromatogr B. 2001;754:97–106.
La S, Cho JH, Kim JH, Kim KR. Capillary electrophoretic profiling and pattern recognition analysis of urinary nucleosides from thyroid cancer patients. Anal Chim Acta. 2003;486:171–82.
Mao Y, Zhao X, Wang S, Cheng Y. Urinary nucleosides based potential biomarker selection by support vector machine for bladder cancer recognition. Anal Chim Acta. 2007;598:34–40.
Zheng YF, Kong HW, Xiong JH, Lv S, Xu GW. Clinical significance and prognostic value of urinary nucleosides in breast cancer patients. Clin Biochem. 2005;38:24–30.
Zheng YF, Yang J, Zhao XJ, Feng B, Kong HW, Chen YJ, et al. Urinary nucleosides as biological markers for patients with colorectal cancer. World J Gastroenterol. 2005;11:3871–6.
Thompson IM, Tangen CM, Kristal AR. Prostate-specific antigen: a misused and maligned prostate cancer biomarker. J Natl Cancer Inst. 2008;100:1487–8.
Thompson IM, Ankerst DP, Chi C, Lucia S, Goodman PJ, Crowley JJ, et al. Operating characteristics of prostate-specific antigen in men with an initial PSA level of 3.0 ng/mL or lower. JAMA. 2005;294:66.
Catalona WJ, Smith DS, Ornstein DK. Prostate cancer detection in men with serum PSA concentrations of 2.6 to 4.0 ng/mL and benign prostate examination. JAMA. 1997;277:1452.
Brawer MK, Meyer GE, Letran JL, Bankson DD, Morris DL, Yeung KK, et al. Measurement of complexed PSA improves specificity for early detection of prostate cancer. Urology. 1998;52:372–8.
Klein EA, Jones JS, editors. Management of prostate cancer. 3rd ed. Humana: New York; 2013.
Mydlo JH, Godec CJ, editors. Prostate cancer: science and clinical practice. 1st ed. Elsevier: London; 2003.
Woolf SH, Jonas S, Lawrence RS, editors. Health promotion and disease prevention in clinical practice. Philadelphia: Lippincott Williams & Wilkins; 1996.
Struck W, Waszczuk-Jankowska M, Kaliszan R, Markuszewski MJ. The state-of-the-art determination of urinary nucleosides using chromatographic techniques “hyphenated” with advanced bioinformatic methods. Anal Bioanal Chem. 2011;401:2039–50.
Simionato AVC, Carrilho E, Maggi Tavares MF. CE-MS and related techniques as a valuable tool in tumor biomarkers research. Electrophoresis. 2010;31:1214–26.
Buzatto AZ, de Sousa AC, Guedes SF, Cieslarová Z, Simionato AVC. Metabolomic investigation of human diseases biomarkers by CE and LC coupled to MS. Electrophoresis. 2014;35:1285–307.
Buzatto AZ, Guedes SF, de Oliveira SM, Gallafrio JM, Simionato AVC. Higher detectability method for the analysis of nucleosides, putative tumor biomarkers, in blood serum samples by CE-UV with reversed EOF. Electrophoresis. 2015;36:2968–75.
Brazilian National Health Surveillance Agency - ANVISA (2003) Guia de validação de métodos bioanalíticos, RE no 899, de 29 de maio de 2003.
Food and Drug Administration (2001) Guidance for industry - bioanalytical method validation. US Department of Health and Human Services, Washington.
European Medicines Agency. Guideline on validation of bioanalytical method validation. London: European Medicines Agency; 2009.
Jiang Y, Ma Y. A fast capillary electrophoresis method for separation and quantification of modified nucleosides in urinary samples. Anal Chem. 2009;81:6474–80.
Szymańska E, Markuszewski MJ, Bodzioch K, Kaliszan R. Development and validation of urinary nucleosides and creatinine assay by capillary electrophoresis with solid phase extraction. J Pharm Biomed Anal. 2007;44:1118–26.
Liebich HM, Xu GW, Di Stefano C, Lehmann R, Häring HU, Lu P, et al. Analysis of normal and modified nucleosides in urine by capillary electrophoresis. Chromatographia. 1997;45:396–401.
Zheng YF, Xu GW, Liu DY, Xiong JH, Zhang PD, Zhang C, et al. Study of urinary nucleosides as biological marker in cancer patients analyzed by micellar electrokinetic capillary chromatography. Electrophoresis. 2002;23:4104–9.
Paz N, Levanon EY, Amariglio N, et al (2007) Altered adenosine-to-inosine RNA editing in human cancer. Genome Res 1586–1595
Grosjean H. Modification and editing of RNA: historical overview and important facts to remember. Top Curr Genet. 2005;12:1–22.
Alseth I, Dalhus B, Bjørås M. Inosine in DNA and RNA. Curr Opin Genet Dev. 2014;26:116–23.
Dominissini D, Moshitch-Moshkovitz S, Amariglio N, Rechavi G. Adenosine-to-inosine RNA editing meets cancer. Carcinogenesis. 2011;32:1569–77.
Gallo A, Locatelli F. ADARs: allies or enemies? The importance of A-to-I RNA editing in human disease: from cancer to HIV-1. Biol Rev Camb Philos Soc. 2012;87:95–110.
Akhavan-Niaki H, Samadani AA. DNA methylation and cancer development: molecular mechanism. Cell Biochem Biophys. 2013;67:501–13.
Ehrlich M. DNA methylation in cancer: too much, but also too little. Oncogene. 2002;21:5400–13.
Carmona FJ, Azuara D, Berenguer-Llergo A, et al. DNA methylation biomarkers for noninvasive diagnosis of colorectal cancer. Cancer Prev Res. 2013;6:656–65.
Formosa A, Lena AM, Markert EK, et al. DNA methylation silences miR-132 in prostate cancer. Oncogene. 2013;32:127–34.
Aran D, Hellman A. DNA Methylation of transcriptional enhancers and cancer predisposition. Cell. 2013;154:11–3.
Aran D, Sabato S, Hellman A. DNA methylation of distal regulatory sites characterizes dysregulation of cancer genes. Genome Biol. 2013;14:R21.
Worley B, Powers R. Multivariate analysis in metabolomics. Curr Metabolomics. 2012;1:92–107.
Ballabio D, Consonni V. Classification tools in chemistry. Part 1: linear models. PLS-DA. Anal. Methods. 2013;5:3790.
Acknowledgments
The authors thank the Clinics Hospital Blood Center (University of Campinas) and Laurione Candido de Oliveira for providing the serum samples from prostate cancer patients, Coordination for the Improvement of Higher Education Personnel (CAPES) for a scholarship to A.Z.B, the National Council for Scientific and Technological Development (CNPq) for financial support, and São Paulo Research Foundation (FAPESP) for financial support and a scholarship for M.O.S..
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare they have no conflict of interest.
Research involving human participants and/or animals
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Declaration of Helsinki and its later amendments or comparable ethical standards.
Ethical approval
This work does not involve any studies with animals performed by any of the authors.
Informed consent
Informed consent was obtained from all individual participants in the study.
Funding
This study was funded by Coordination for the Improvement of Higher Education Personnel (CAPES), the National Council for Scientific and Technological Development (CNPq), and São Paulo Research Foundation (FAPESP).
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 468 kb)
Rights and permissions
About this article
Cite this article
Buzatto, A.Z., de Oliveira Silva, M., Poppi, R.J. et al. Assessment of nucleosides as putative tumor biomarkers in prostate cancer screening by CE–UV. Anal Bioanal Chem 409, 3289–3297 (2017). https://doi.org/10.1007/s00216-017-0297-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-017-0297-7