Abstract
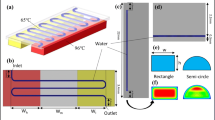
A parallel-processing four-station polymerase chain reaction (PCR) device has been developed, which performs continuous-flow PCR without optimization of the annealing temperature. Since the annealing temperature of each station can be controlled independently, the device covers an annealing temperature range of 50–68 °C, which is wide enough to perform PCR for any DNA fragment regardless of its optimum annealing condition. This arrangement lets us continuously obtain an amplified amount of a DNA fragment at least from one of the stations. The device consists of four identical cylindrical stations (diameter 20 mm, height 55 mm). A polytetrafluoroethylene capillary reactor (length 2 m, I.D. 100 μm, O.D. 400 μm) is wound helically up around each station. The whole assembly is designed to minimize the number of heating blocks (for providing temperatures of denaturation, annealing, and extension) to be seven and to shape a compact cube (height 55 mm, base 60 mm × 60 mm). The reproducibility for continuous-flow PCR is reasonably high (run-to-run and station-to-station relative standard deviation of their amplification is lower than 6 % and about 4 %, respectively). Performance on the optimization-free DNA amplification has been evaluated with four DNA samples with different annealing conditions and product sizes (323, 608, 828, and 1101 bp), which has demonstrated that in all cases, PCR is successful at least on one station. In addition, three DNA fragments with different lengths (323, 1101, and 2836 bp) have been successfully amplified in a segmented-flow mode without the carry-over contamination between segments. This result suggests that this device could serve as the PCR module of a continuous-flow high-throughput on-line total DNA analysis system integrating all necessary modules from cell lysis/DNA extraction to PCR product analysis.





Similar content being viewed by others
References
Saiki RK, Scharf S, Faloona F, Mullis KB, Horn GT, Erlich HA, et al. Enzymatic amplification of β-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science. 1985;230:1350–4.
Jung JH, Choi SJ, Park BH, Choi YK, Seo TS. Ultrafast rotary PCR system for multiple influenza viral RNA detection. Lab Chip. 2012;12:1598–600.
Butler JM. Forensic STR typing: biology, technology and genetics of STR markers. 2nd ed. New York: Elsevier; 2005.
Liu P, Seo TS, Beyor N, Shin K-J, Scherer JR, Mathies RA. Integrated portable polymerase chain reaction-capillary electrophoresis microsystem for rapid forensic short tandem repeat typing. Anal Chem. 2007;79:1881–9.
Regan JF, Makarewicz AJ, Hindson BJ, Metz TR, Gutierrez DM, Corzett TH, et al. Environmental monitoring for biological threat agents using the autonomous pathogen detection system with multiplexed polymerase chain reaction. Anal Chem. 2008;80:7422–9.
Zhang Y, Ozdemir P. Microfluidic DNA amplification–a review. Anal Chim Acta. 2009;638:115–25.
Zhang Y, Jiang H-R. A review on continuous-flow microfluidic PCR in droplets: advances, challenges and future. Anal Chim Acta. 2016;914:7–16.
Phaneuf CR, Pak N, Forest CR. Modeling radiative heating of liquids in microchip reaction chambers. Sensors Actuators A. 2011;167:531–6.
Amasia M, Cozzens M, Madou MJ. Centrifugal microfluidic platform for rapid PCR amplification using integrated thermoelectric heating and ice-valving. Sensors Actuators B. 2012;161:1191–7.
Sugumar D, Kong LX, Ismail A, Ravichandran M, Yin LS. Rapid multi sample DNA amplification using rotary-linear polymerase chain reaction device (PCRDisc). Biomicrofluidics. 2012;6:014119.
Zhang R, Gong H-Q, Zeng X, Lou CP, Sze CC. A microfluidic liquid phase nucleic acid purification chip to selectively isolate DNA or RNA from low copy/single bacterial cells in minute sample volume followed by direct on-chip quantitative PCR assay. Anal Chem. 2013;85:1484–91.
Houssin T, Cramer J, Grojsman R, Bellahsene L, Colas G, Moulet H, et al. Ultrafast, sensitive and large-volume on-chip real-time PCR for the molecular diagnosis of bacterial and viral infections. Lab Chip. 2016;16:1401–11.
Hartung R, Brösing A, Sczcepankiewicz G, Liebert U, Häfner N, Dürst M, et al. Application of an asymmetric helical tube reactor for fast identification of gene transcripts of pathogenic viruses by micro flow-through PCR. Biomed Microdevices. 2009;11:685–92.
Peham JR, Grienauer W, Steiner H, Heer R, Vellekoop MJ, Nöhammer C, et al. Long target droplet polymerase chain reaction with a microfluidic device for high-throughput detection of pathogenic bacteria at clinical sensitivity. Biomed Microdevices. 2011;13:463–73.
Wu W, Lee NY. Three-dimensional on-chip continuous-flow polymerase chain reaction employing a single heater. Anal Bioanal Chem. 2011;400:2053–60.
Chung KH, Choi YH, Park S. Development of a continuous-flow polymerase chain reaction device utilizing a polymer disk with a spiral microchannel of gradually varying width. Sensors Actuators B. 2014;191:75–85.
Moschou D, Vourdas N, Kokkoris G, Papadakis G, Parthenios J, Chatzandroulis S, et al. All-plastic, low-power, disposable, continuous-flow PCR chip with integrated microheaters for rapid DNA amplification. Sensors Actuators B. 2014;199:470–8.
Shu B, Zhang C, Xing D. Segmented continuous-flow multiplex polymerase chain reaction microfluidics for high-throughput and rapid foodborne pathogen detection. Anal Chim Acta. 2014;826:51–60.
Tachibana H, Saito M, Tsuji K, Yamanaka K, Hoa LQ, Tamiya E. Self-propelled continuous-flow PCR in capillary-driven microfluidic device: microfluidic behavior and DNA amplification. Sensors Actuators B. 2015;206:303–10.
Liu P, Li X, Greenspoon SA, Scherer JR, Mathies RA. Integrated DNA purification, PCR, sample cleanup, and capillary electrophoresis microchip for forensic human identification. Lab Chip. 2011;11:1041–8.
Jha SK, Chand R, Han D, Jang Y-C, Ra G-S, Kim JS, et al. An integrated PCR microfluidic chip incorporating aseptic electrochemical cell lysis and capillary electrophoresis amperometric DNA detection for rapid and quantitative genetic analysis. Lab Chip. 2012;12:4455–64.
Jiang X, Jing W, Zheng L, Liu S, Wu W, Sui G. A continuous-flow high-throughput microfluidic device for airborne bacteria PCR detection. Lab Chip. 2014;14:671–6.
Rychlik W, Spencer WJ, Rhoads RE. Optimization of the annealing temperature for DNA amplification in vitro. Nucleic Acids Res. 1990;18:6409–12.
Zhang C, Xing D. Microfluidic gradient PCR (MG-PCR): a new method for microfluidic DNA amplification. Biomed Microdevices. 2010;12:1–12.
Park N, Kim S, Hahn JH. Cylindrical compact thermal-cycling device for continuous-flow polymerase chain reaction. Anal Chem. 2003;75:6029–33.
Fuchiwaki Y, Saito M, Wakida S-I, Tamiya E, Nagai H. A practical liquid plug flow-through polymerase chain-reaction system based on a heat-resistant resin chip. Anal Sci. 2011;27:225–30.
Njoroge SK, Witek MA, Battle KN, Immethun VE, Hupert ML, Soper SA. Integrated continuous flow polymerase chain reaction and micro-capillary electrophoresis system with bioaffinity preconcentration. Electrophoresis. 2011;32:3221–32.
Zhang P, Nan H, Lee M-J, Kang SH. Ultra-fast separation of infectious disease-related small DNA molecules by single- and multi-channel microchip electrophoresis. Talanta. 2013;106:388–93.
Wittwer CT, Garling DJ. Rapid cycle DNA amplification: time and temperature optimization. BioTechniques. 1991;10:76–83.
Li Y, Zhang C, Xing D. Fast identification of foodborne pathogenic viruses using continuous-flow reverse transcription-PCR with fluorescence detection. Microfluid Nanofluid. 2011;10:367–80.
Sugumar D, Ismail A, Ravichandran M, Aziah I, Kong LX. Amplification of SPPS150 and Salmonella typhi DNA with a high throughput oscillating flow polymerase chain reaction device. Biomicrofluidics. 2010;4:024103.
Shaw KJ, Joyce DA, Docker PT, Dyer CE, Greenway GM, Greenman J, et al. Development of a real-world direct interface for integrated DNA extraction and amplification in a microfluidic device. Lab Chip. 2011;11:443–8.
Wu W, Kang K-T, Lee NY. Bubble-free on-chip continuous-flow polymerase chain reaction: concept and application. Analyst. 2011;136:2287–93.
Chen JJ, Shen CM, Ko YW. Analytical study of a microfludic DNA amplification chip using water cooling effect. Biomed Microdevices. 2013;15:261–78.
Trinh KTL, Wu W, Lee NY. Planar poly(dimethylsiloxane) (PDMS)–glass hybrid microdevice for a flow-through polymerase chain reaction (PCR) employing a single heater assisted by an intermediate metal alloy layer for temperature gradient formation. Sensors Actuators B. 2014;190:177–84.
Chen JJ, Liao MH, Li KT, Shen CM. One-heater flow-through polymerase chain reaction device by heat pipes cooling. Biomicrofluidics. 2015;9:014107.
Lim K, Kim S, Hahn JH. Roller-type squeezing pump with picoliter handling capability. Sensors Actuators B. 2003;92:208–14.
Ha JW, Hahn JH. Acupuncture sample injection for microchip capillary electrophoresis and electrokinetic chromatography. Anal Chem. 2016;88:4629–34.
Hahn JH, Kwak BJ, Kim H-O, Microchip for analyzing nucleic acid and method of nucleic acid analysis using the same. Patent 9,017,941. 2015.
Kim H-O, Kwak B-J, Hahn JH. Compact continuous-flow PCR system and on-line DNA analysis. Orlando: Pittsburgh Conference on Analytical Chemistry and Applied Spectroscopy 2012: Proceedings of the PITTCON 2012 Symposium; 2012. 11–15 March 2012 (PITTCON 2012).
Acknowledgments
This research was supported by the R&D Center for Green Patrol Technologies through the R&D for Global Top Environmental Technologies (RE201604086) funded by the Ministry of Environment, Republic of Korea (MOE). This work was also supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIP) (NRF-2015R1A2A2A04005445).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 152 kb)
Rights and permissions
About this article
Cite this article
Kim, H., Park, N. & Hahn, J.H. Parallel-processing continuous-flow device for optimization-free polymerase chain reaction. Anal Bioanal Chem 408, 6751–6758 (2016). https://doi.org/10.1007/s00216-016-9798-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-016-9798-z