Abstract
A single-probe strip test for the rapid and sensitive detection of miRNA-21 mimics is reported herein. Highly specific structurally responsive bi-functional, thiol and biotin, DNA/LNA oligonucleotide probes (molecular beacons-MB) were designed and conjugated with gold nanoparticles (AuNPs) (i.e. biotin-MB-AuNPs). The proposed design had the ability to modulate the accessibility of the biotin group as a function of the presence of a miRNA target allowing the interaction of the boilable with the streptavidin test zone only in the presence of the miRNA-21 mimics. For quantitative evaluation, images of the strip tests were recorded using a flatbed scanner (Epson Perfection V370 Photo). The colour intensities of the test zones of the strip tests were analysed with the ImageJ software (Scion Corp., USA) and quantified as a function of pixel intensity. The response of the strip test was linear over the range 0.5 to 20 nM miRNA-21 (limit of detection of 115 pM) and showed good reproducibility (intra and inter CVs below 8 %); furthermore, the assay was shown to be highly selective, discriminating other interference miRNAs mimics (e.g. miRNA-221 and miRNA-205). Finally, the proposed strip test was used for detection of miRNA-21 mimics in spiked serum samples, demonstrating its potential for point-of-care clinical applications. Main advantages of the single-probe strip test design are its versatility, simplicity and robustness, which can be easily extended to other miRNA targets by tuning the sequence of the single probe. Furthermore, the use of the structurally responsive single probe is particularly relevant in the case of short-length targets, such as miRNA, whereas a conventional sandwich approach might require a careful control of assay conditions such as hybridization temperature and salt concentration.

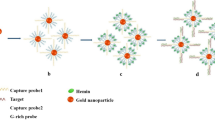
Pictorial representation of the working principle of the proposed structurally responsive oligonucleotide-based single-probe lateral-flow test and example of the test response as a function of increasing concentration of target analyte.





Similar content being viewed by others
References
Zhou Y, Zhang Z, Xu Z, Yin H, Ai S. MicroRNA-21 detection based on molecular switching by amperometry. New J Chem. 2012;36(10):1985–91.
Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294(5543):853–8.
Small EM, Frost RJA, Olson EN. MicroRNAs add a new dimension to cardiovascular disease. Circulation. 2010;121(8):1022–32.
Lu Z, Liu M, Stribinskis V, Klinge CM, Ramos KS, Colburn NH, et al. MicroRNA-21 promotes cell transformation by targeting the programmed cell death 4 gene. Oncogene. 2008;27(31):4373–9.
Asaga S, Kuo C, Nguyen T, Terpenning M, Giuliano AE, Hoon DSB. Direct serum assay for microRNA-21 concentrations in early and advanced breast cancer. Clin Chem. 2011;57(1):84–91.
Markou A, Tsaroucha EG, Kaklamanis L, Fotinou M, Georgoulias V, Lianidou ES. Prognostic value of mature microRNA-21 and microRNA-205 overexpression in non–small cell lung cancer by quantitative real-time RT-PCR. Clin Chem. 2008;54(10):1696–704.
Wei J, Gao W, Zhu C-J, Liu Y-Q, Mei Z, Cheng T, et al. Identification of plasma microRNA-21 as a biomarker for early detection and chemosensitivity of non–small cell lung cancer. Chin J Cancer. 2011;30(6):407–14.
Zhang HL, Yang LF, Zhu Y, Yao XD, Zhang SL, Dai B, et al. Serum miRNA-21: elevated levels in patients with metastatic hormone-refractory prostate cancer and potential predictive factor for the efficacy of docetaxel-based chemotherapy. Prostate. 2011;71(3):326–31.
Kim SW, Li Z, Moore PS, Monaghan AP, Chang Y, Nichols M, et al. A sensitive non-radioactive northern blot method to detect small RNAs. Nucleic Acids Res. 2010;38(7), e98.
Cissell KA, Rahimi Y, Shrestha S, Hunt EA, Deo SK. Bioluminescence-based detection of microRNA, miR21 in breast cancer cells. Anal Chem. 2008;80(7):2319–25.
Wang ZX, Bian HB, Wang JR, Cheng ZX, Wang KM, De W, et al. Prognostic significance of serum miRNA-21 expression in human non-small cell lung cancer. Oncology. 2011;104(7):847–51.
Yin H, Zhou Y, Zhang H, Meng X, Ai S. Electrochemical determination of microRNA-21 based on graphene, LNA integrated molecular beacon, AuNPs and biotin multifunctional bio bar codes and enzymatic assay system. Biosens Bioelectron. 2012;33(1):247–53.
Li W, Ruan K. MicroRNA detection by microarray. Anal Bioanal Chem. 2009;394(4):1117–24.
Hou SY, Hsiao YL, Lin MS, Yen CC, Chang CS. MicroRNA detection using lateral flow nucleic acid strips with gold nanoparticles. Talanta. 2012;99:375–9.
Gao X, Xu H, Baloda M, Gurung AS, Xu LP, Wang T, et al. Visual detection of microRNA with lateral flow nucleic acid biosensor. Biosens Bioelectron. 2014;54:578–84.
Fang X, Liu X, Schuster S, Tan W. Designing a novel molecular beacon for surface-immobilized DNA hybridization studies. J Am Chem Soc. 1999;121(12):2921–2.
Tyagi S, Kramer FR. Molecular beacons: probes that fluoresce upon hybridization. Nat Biotechnol. 1996;14(3):303–8.
Beni V, Hayes K, Lerga TM, O’Sullivan CK. Development of a gold nano-particle-based fluorescent molecular beacon for detection of cystic fibrosis associated mutation. Biosens Bioelectron. 2010;26(2):307–13.
Nasef H, Beni V, O’Sullivan CK. Electrochemical molecular beacon DNA biosensor for the detection and discrimination of the DF508 cystic fibrosis mutation. J Electroanal Chem. 2011;662(2):322–7.
Alexander JC, Pandit A, Bao G, Connolly D, Rochev Y. Monitoring mRNA in living cells in a 3D in vitro model using TAT-peptide linked molecular beacons. Lab Chip. 2011;11(22):3908–14.
Bratu DP, Catrina IE, Marras SAE. Tiny molecular beacons for in vivo mRNA detection. In: Gerst JE, Editors. RNA Detection and Visualization. Humana Press; 2011. p. 141–157.
Catrina IE, Marras SAE, Bratu DP. Tiny molecular beacons: LNA/2′-O-methyl RNA chimeric probes for imaging dynamic mRNA processes in living cells. ACS Chem Biol. 2012;7(9):1586–95.
Chen AK, Behlke MA, Tsourkas A. Avoiding false-positive signals with nucleasevulnerable molecular beacons in single living cells. Nucleic Acids Res. 2007;35(16), e105.
van den Bogaard PC, Tyagi S. Using molecular beacons to study dispersal of mRNPs from the gene locus. In: Hancock R, editors. The Nucleus. Humana Press; 2008. p. 91–103.
Alsmadi O, Al-Rubeaan K, Wakil SM, Imtiaz F, Mohamed G, Al-Saud H, et al. Genetic study of Saudi diabetes (GSSD): significant association of the KCNJ11 E23K polymorphism with type 2 diabetes. Diabetes Metab Res Rev. 2008;24(2):137–40.
Bhagwat A, Patel J, Chua T, Chan A, Cruz S, Aguilar GG. Detection of Salmonella species in foodstuffs. In: Marx A, Seitz O, editors. Molecular beacons: signalling nucleic acid probes, methods, and protocols. Humana Press; 2008. p. 33–43.
Blackard JT, Komurian-Pradel F, Perret M, Sodoyer M, Smeaton L, Clair JBS, et al. Intrahepatic cytokine expression is downregulated during HCV/HIV co‐infection. J Med Virol. 2006;78(2):202–7.
Chen W, Martinez G, Mulchandani A. Molecular beacons: a real-time polymerase chain reaction assay for detecting Salmonella. Anal Biochem. 2000;280(1):166–72.
Hadjinicolaou A, Farcas G, Demetriou V, Mazzulli T, Poutanen S, Willey B, et al. Development of a molecular-beacon-based multi-allelic real-time RT-PCR assay for the detection of human coronavirus causing severe acute respiratory syndrome (SARS-CoV): a general methodology for detecting rapidly mutating viruses. Arch Virol. 2011;156(4):671–80.
Culha M, Stokes DL, Griffin GD, Vo-Dinh T. Application of a miniature biochip using the molecular beacon probe in breast cancer gene BRCA1 detection. Biosens Bioelectron. 2004;19(9):1007–12.
Epstein JR, Leung APK, Lee K-H, Walt DR. High-density, microsphere-based fiber optic DNA microarrays. Biosens Bioelectron. 2003;18(5–6):541–6.
Kim H, Kane MD, Kim S, Dominguez W, Applegate BM, Savikhin S. A molecular beacon DNA microarray system for rapid detection of E. coli O157: H7 that eliminates the risk of a false negative signal. Biosens Bioelectron. 2007;22(6):1041–7.
Martinez K, Estevez MC, Wu Y, Phillips JA, Medley CD, Tan W. Locked nucleic acid based beacons for surface interaction studies and biosensor development. Anal Chem. 2009;81(9):3448–54.
El-Yazbi AF, Loppnow GR. Detecting UV-induced nucleic-acid damage. TrAC Trends Anal Chem. 2014;61:83–91.
Baker MB, Bao G, Searles CD. In vitro quantification of specific microRNA using molecular beacons. Nucleic Acids Res. 2012;40(2), e13.
Mao X, Xu H, Zeng Q, Zeng L, Liu G. Molecular beacon-functionalized gold nanoparticles as probes in dry-reagent strip biosensor for DNA analysis. Chem Commun. 2009;(21): 3065–3067.
Guo Z, Duan J, Yang F, Li M, Hao T, Wang S, et al. A test strip platform based on DNA-functionalized gold nanoparticles for on-site detection of mercury (II) ions. Talanta. 2012;93:49–54.
Xu H, Mao X, Zeng Q, Wang S, Kawde A-N, Liu G. Aptamer-functionalized gold nanoparticles as probes in a dry-reagent strip biosensor for protein analysis. Anal Chem. 2008;81(2):669–75.
He Y, Zeng K, Gurung AS, Baloda M, Xu H, Zhang X, et al. Visual detection of single-nucleotide polymorphism with hairpin oligonucleotide-functionalized gold nanoparticles. Anal Chem. 2010;82(17):7169–77.
He Y, Zhang X, Zhang S, Kris MK, Man FC, Kawde AN, et al. Visual detection of single-base mismatches in DNA using hairpin oligonucleotide with double-target DNA binding sequences and gold nanoparticles. Biosens Bioelectron. 2012;34(1):37–43.
Meng X, Xu M, Zhu J, Yin H, Ai S. Fabrication of DNA electrochemical biosensor based on gold nanoparticles, locked nucleic acid modified hairpin DNA and enzymatic signal amplification. Electrochim Acta. 2012;71:233–8.
Greene SB, Herschkowitz JI, Rosen JM. The ups and downs of miR-205: identifying the roles of miR-205 in mammary gland development and breast cancer. RNA Biol. 2010;7(3):300–4.
Majid S, Dar AA, Saini S, Yamamura S, Hirata H, Tanaka Y, et al. MicroRNA‐205–directed transcriptional activation of tumor suppressor genes in prostate cancer. Cancer. 2010;116(24):5637–49.
Wiklund ED, Bramsen JB, Hulf T, Dyrskjøt L, Ramanathan R, Hansen TB, et al. Coordinated epigenetic repression of the miR‐200 family and miR‐205 in invasive bladder cancer. Int J Cancer. 2011;128(6):1327–34.
Lu Q, Lu C, Zhou G-P, Zhang W, Xiao H, Wang X-R. MicroRNA-221 silencing predisposed human bladder cancer cells to undergo apoptosis induced by TRAIL. Urol Oncol: Sem Orig Investig. 2010;28(6):635–41.
Conti A, Aguennouz MH, La Torre D, Tomasello C, Cardali S, Angileri F, et al. miR-21 and 221 upregulation and miR-181b downregulation in human grade II–IV astrocytic tumors. J Neurooncol. 2009;93(3):325–32.
Liu J, Lu Y. Preparation of aptamer-linked gold nanoparticle purple aggregates for colorimetric sensing of analytes. Nat Protocols. 2006;1(1):246–52.
Lesnik EA, Freier SM. Relative thermodynamic stability of DNA, RNA, and DNA:RNA hybrid duplexes: relationship with base composition and structure. Biochemistry. 1995;34(34):10807–15.
Lang BE, Schwarz FP. Thermodynamic dependence of DNA/DNA and DNA/RNA hybridization reactions on temperature and ionic strength. Biophys Chem. 2007;131(1–3):96–104.
Beni V, Zewdu T, Joda H, Katakis I, O’Sullivan CK. Gold nanoparticle fluorescent molecular beacon for low-resolution DQ2 gene HLA typing. Anal Bioanal Chem. 2012;402(3):1001–9.
Chen T, Wu CS, Jimenez E, Zhu Z, Dajac JG, You M, et al. DNA micelle flares for intracellular mRNA imaging and gene therapy. Angew Chem. 2013;125:2066–70.
Hurst SJ, Lytton-Jean AKR, Mirkin CA. Maximizing DNA loading on a range of gold nanoparticle sizes. Anal Chem. 2006;78(24):8313–8.
Pritchard CC, Cheng HH, Tewari M. MicroRNA profiling: approaches and considerations. Nat Rev Genet. 2012;13(5):358–69.
Acknowledgments
Kamalodin Kor is grateful to the Ministry of Science, Research and Technology of Iran for financial support (project ref. 214140) to carry out this project at Linköping University, Sweden.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflicting interests.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 2020 kb)
Rights and permissions
About this article
Cite this article
Kor, K., Turner, A.P.F., Zarei, K. et al. Structurally responsive oligonucleotide-based single-probe lateral-flow test for detection of miRNA-21 mimics. Anal Bioanal Chem 408, 1475–1485 (2016). https://doi.org/10.1007/s00216-015-9250-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-015-9250-9