Abstract
One of the main problems of anti-cancer therapy is an insufficient differentiation between normal and malignant cells by the known anti-proliferant agents. The antibody-directed enzyme prodrug therapy is a promising approach for a selective treatment of cancer, in which a non-toxic prodrug is enzymatically converted into a highly cytotoxic drug at the surface of malignant cells by a targeted antibody–enzyme conjugate. The transformations and the stability of a very promising novel prodrug and its corresponding cytotoxic derivative were now investigated in detail by high-performance liquid chromatography (HPLC)–mass spectrometry (MS). In order to determine the time-dependent DNA alkylation efficiency and the sequence selectivity of the novel compounds, DNA binding studies using direct electrospray–Fourier transform ion cyclotron resonance–MS (ESI–FTICR–MS) have been performed. These measurements were accompanied by HPLC analyses followed by MS of the separated species to confirm the results of the direct ESI–FTICR–MS measurements. The sites of DNA alkylation could be identified unambiguously by the mass spectrometric fragmentation pattern of the alkylated oligodeoxynucleotides as well as by the results of HPLC followed by MS. A combination of all techniques applied led to a better understanding of the mode of action of the new therapeutics and might be used for an estimation of the cytotoxicity of different prodrugs and drugs since the alkylation efficiency correlates with the bioactivity of the compounds in cell culture investigations.

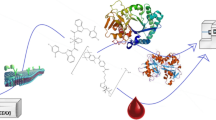
After enzymatic cleavage of the sugar moiety, the untoxic prodrug is converted rapidly into the corresponding highly cytotoxic drug that alkylates DNA with high efficiency
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Despite considerable efforts towards the development of efficient and selective cancer therapies, most anti-cancer agents applied so far in the clinic cause severe side effects because their cytotoxic action is not limited to the tumour cells. To minimise these side effects, current research focuses on the development of targeted tumour therapies. In antibody-directed enzyme prodrug therapy (ADEPT), relatively non-toxic prodrugs are converted to their corresponding highly cytotoxic drugs selectively in the tumour tissue by targeted antibody–enzyme conjugates [1-3]. The toxic action of the drugs is thus limited mainly to the tumour site. For a successful application of ADEPT, water-soluble and stable prodrugs with low cytotoxicity that yield highly cytotoxic drugs after enzymatic activation are needed [4]. Furthermore, insights into the mode of action of the potential therapeutics are highly desirable for a safe drug administration and a sophisticated drug development.
Recently, we have prepared a series of novel glycosidic prodrugs like the anti-methyl-seco-CBI-DMAI-galactoside 1, which can be activated enzymatically by cleavage of the sugar moiety using commercially available β-d-galactosidase from Escherichia coli to give the corresponding seco-drug 2 (Fig. 1) [5, 6]. 2 was expected to cyclise rapidly under loss of HCl to produce the active drug 3 which is an analogue of the highly cytotoxic natural antibiotic CC-1065 (4) [7] and the duocarmycins (e.g. duocarmycin SA (5)) [8].
Galactosidic prodrug 1 containing a detoxifying sugar moiety, enzymatic activation of prodrug 1 to give seco-drug 2 followed by a cyclisation of 2 to afford the active drug 3 as an analogue of the natural products CC-1065 (4) and duocarmycin SA (5). Inactivation of 1, 2 or 3 by hydrolysis leads to 6 and 7, respectively
Since 4 and 5 are supposed to exert their cytotoxic action through alkylation of adenine in AT-rich minor grooves of double-stranded DNA, drugs such as 3 are expected to alkylate cellular DNA, and it can be assumed that the sequence selectivity in the DNA alkylation correlates to the biological activity [9-11]. By means of cell culture investigations using human bronchial carcinoma cells of line A549, it was shown that prodrug 1 is 4,800 times less toxic than its corresponding drug 3, which exhibits a high cytotoxicity with an IC50 of 0.75 nM [5, 6].
Here, we describe investigations aimed at a better understanding of the mode of action of prodrugs such as 1. First, the stability of prodrug 1 and of the enzymatic activation of the prodrug to give the corresponding seco-drug 2 using high-performance liquid chromatography (HPLC)–mass spectrometry (MS) was investigated. Furthermore, the transformations of the seco-drug 2 to the drug 3 under cell culture conditions and of the further transformations of the drug were studied by means of HPLC–MS. Second, the time-dependent DNA alkylation efficiency and the sequence selectivity of prodrug 1 and of its corresponding seco-drug 2 were investigated using HPLC followed by MS as well as by means of direct electrospray ionisation (ESI)–Fourier transform ion cyclotron resonance (FTICR)–MS. A short communication of parts of this work, namely the use of direct ESI–FTICR–MS for the investigation of the alkylation reaction, has recently been published [12].
Experimental
Materials
Prodrug 1 and seco-drug 2 were synthesised according to previously published procedures [5] and stock solutions in dimethyl sulfoxide (DMSO) were prepared. UltraCulture® (Lonza, Germany) supplemented with 2 mm l-glutamine from Gibco was used as cell culture medium. For the investigation of the enzymatic activation of 1, β-d-galactosidase from E. coli (E.C. 3.2.1.23, G 5635, activity 700 units (U) per milligramme protein at pH 7.3 and 37 °C, 1 U = conversion of 1 µmol of substrate per minute, Sigma, Germany) was added. The synthetic dsDNA oligomer 5′-d(CGGTCAATTAGTCGC)-3′ (ON-1) ∙ 3′-d(GCCAGTTAATCAGCG)-5′ (ON-2) was purchased from IBA (Göttingen, Germany) as aqueous solution (0.1 mm) of the respective sodium salt.
Methods
Transformations of prodrug 1 and seco-drug 2 in cell culture medium
Ten microlitres of a stock solution of 1 or 2 in DMSO (48 mm) was mixed with 990 µL cell culture medium, and the resulting reaction mixture was incubated at 37 °C for 24 h. Samples were taken after 0.0, 0.5, 1.0, 1.5, 2.0, 3.0, 4.0, 5.0, 6.0 and 24.0 h. For the investigation of the enzymatic activation of 1, β-d-galactosidase (10 U mL−1) was added prior to the incubation and additional samples were taken after 2.5 and 7 h. The samples without β-d-galactosidase were directly investigated by HPLC–MS. The samples with β-d-galactosidase were subjected to ultracentrifugation using common polymerase chain reaction (PCR) filters (UFC7PCR50, Millipore, Germany) to remove the enzyme; the filter was washed with an equivalent amount of methanol, and HPLC–MS of the combined eluents was performed.
High-performance liquid chromatography–mass spectrometry
Chromatography was performed with an HPLC system consisting of a Rheos 4000 solvent pump and an ERC-3415 degasser from Flux Instruments, an autosampler 851-AS from Jasco and a diode array detector Surveyor PDA operated at 200–800 nm from Thermo. As column, a Synergi Max-RP C12 (150 × 2 mm, 4 µm) from phenomenex was used. Samples were eluted within 15 min with a flow rate of 300 µL min−1 by applying a gradient (0–15 min 30→100% B, 15–22 min 100% B, 22–23 min 100→30% B, 23–29 min 30% B). Eluent A was water with 0.05% formic acid from Roth. Eluent B was methanol from VWR with 0.05% formic acid from Roth. Online ESI mass spectrometry was performed in the positive mode with an ion-trap mass spectrometer LCQ from Finnigan with detection in the mass range of 100–2000 m/z. The capillary temperature was set to 220 °C, the spray voltage to 4.5 kV and the sheath gas flow to 80 (arbitrary units). For each measurement, the area under the curve (AuC, reconstructed ion chromatogram versus retention time) of all species including an internal standard was determined and the respective relative area under the curve was calculated. The measurements were performed in triplicate, and the median relative area under the curve of each species was then plotted versus time.
Investigation of the DNA alkylation of prodrug 1 and seco-drug 2
The reactions were carried out in aqueous solution with 0.1 mmol L−1 dsDNA and prodrug 1 or seco-drug 2 in a 1:1 and 1:5 ratio. One microlitre of stock solution of 1 or 2 was mixed with 100 µL of dsDNA in water, and the reaction mixture was incubated at 25 °C for 24 h. For direct investigations of the reaction mixtures, samples were taken at 0 and 24 h, diluted with an equivalent amount of methanol and the resulting solution introduced directly into the ion source of the ESI–FTICR mass spectrometer. For comparison, the samples were separated using HPLC, the sample fractions lyophilised, dissolved in a mixture of water/methanol (1:1) and the resulting solutions introduced into the ion source of the ESI–FTICR mass spectrometer. Additionally, selected samples were heated to 95 °C for 2 h and then subjected to HPLC, lyophilisation and high-resolution mass spectrometry.
Semi-preparative high-performance liquid chromatography
HPLC separations were performed with an Agilent 1200 with diode array detector from Agilent Technologies, an Aquapore OD-300 Column (220 × 4.6 mm, 7 µm) from Perkin Elmer and a Bondapak® C18 Column (300 × 3.9 mm, particle size 10 µm, pore size 125 Å) from Waters. Samples were eluted within 45 min with a flow rate of 1 mL min−1 (Aquapore OD-300) or 2 mL min−1 (Bondapak® C18) at 28 °C by applying a two-stage gradient (0–2 min 5% B, 2–22 min 5→20% B, 55–45 min 20→80% B, 45–50 min 80% B, 50–60 min 80→5% B). Eluent A was 0.1 mol L−1 triethylammonium acetate buffer (H2O, pH 7.0). Eluent B was 0.1 mol L−1 triethylammonium acetate buffer (80% acetonitrile and 20% water, pH 7.0).
High-resolution electrospray mass spectrometry
High-resolution mass spectrometry was performed using a 7-T FTICR–MS instrument (APEX IV, Bruker Daltonics, Billerica, USA) equipped with an APOLLO electrospray ion source and a syringe pump (74900 series, Cole-Parmer, Vernon Hills, USA) with a flow rate of 2 µL min−1 for sample injection. The ions generated in the negative ion mode were accumulated in the hexapole region for 0.8 s and transferred subsequently into the ion cyclotron resonance (ICR) cell. For gentle desolvation, the drying gas temperature was set to 100 °C and the capillary exit voltage to −100 V. Enh anced fragmentation of alkylated oligonucleotides was achieved by capillary-skimmer dissociation (CSD) with a capillary exit voltage of −150 V. Collision-induced dissociation (CID)–MS/MS measurements were carried out by fragmentation of ions isolated in the ICR cell using argon as collision gas.
Results and discussion
First, the stability of prodrug 1 in cell culture medium at 37 °C was determined using HPLC–MS (Fig. 2, for mass spectra, tables and chromatograms: see Supplementary Material Figs. S1–S2 and Table S1). In the time period of 24 h, no cleavage of the sugar moiety in 1, i.e. no activation of 1 to give the corresponding highly cytotoxic derivatives 2 and 3, was observed. Instead, as was obvious from the chromatograms and the respective mass spectra, nucleophilic substitution of the chlorine atom in 1 by a hydroxyl group occurred, transforming 1 into to the derivative 6. In the absence of β-d-galactosidase, 1 has a half-life of approximately 14 ± 1 hours.
In the presence of β-d-galactosidase, prodrug 1 was efficiently activated to the corresponding seco-drug 2 by removal of the sugar moiety in 1 (Fig. 3, for additional chromatograms and tables: see Supplementary Material Fig. S3 and Table S2).
Transformation of prodrug 1 in the presence of β-d-galactosidase to the corresponding seco-drug 2 and rapid cyclisation of 2 to the drug 3. Hydrolysis of 1 and 3 to the hydroxylated species 6 and 7, respectively, and formation of a dimeric drug. AuC against time and chromatograms after 0.0 h and 1.5 h of incubation
As was expected, 2 cyclised rapidly under loss of HCl to give the highly reactive drug 3 directly after its generation from the prodrug 1. More slowly, prodrug 1 and drug 3 were both hydrolysed to provide the hydroxylated species 6 and 7, respectively. Furthermore, in the absence of other potential nucleophiles, drug 3 very slowly reacted to give a dimeric species.
In order to investigate the kinetics of the cyclisation of seco-drug 2 to the corresponding drug 3 in detail, the transformations of 2 in cell culture medium at 37 °C were studied using the same method as described for the prodrug 1 (Fig. 4, for mass spectra, chromatograms and tables: see Supplementary Material Figs. S4–S5 and Table S3).
As was already observed for the seco-drug 2 that is generated from 1 in situ (Fig. 3), a pure sample of 2 cyclises rapidly to give the corresponding drug 3 after its mixing with cell culture medium (Fig. 4). 3 is then hydroxylated by nucleophilic substitution and reacts to a dimeric derivative. The identity of each compound was verified by ESI–MS and ESI–MS/MS experiments. Thus, it was shown that the dimeric species is formed by a reaction of the hydroxylated drug 7 with the drug 3 (for MS and MS/MS spectra: see Supplementary Material Fig. S5). A dimerisation of the drug under physiological conditions is unlikely to occur due to the presence of a multitude of nucleophiles with higher reactivity in biological systems and the lower concentrations used for a therapeutic application as compared to the HPLC–MS studies. The deactivation of the drug 3 in the time window observed is desirable for a clinical application of prodrug 1 because a long lifetime of 3 might cause undesirable side effects due to its leaking out of the tumour site by means of diffusion.
In order to investigate the DNA alkylation efficiency of prodrug 1 and seco-drug 2, both compounds were incubated with the synthetic double-stranded DNA oligomer 5′-d(CGGTCAATTAGTCGC)-3′ (ON-1) ∙ 3′-d(GCCAGTTAATCAGCG)-5′ (ON-2) in aqueous solution [12].
After incubation of the dsDNA ON-1∙ON-2 with the seco-drug 2 in a ratio of 1:1 for 24 h, a very efficient and selective alkylation of the oligonucleotide ON-1 by the intermediately formed drug 3 could be observed using direct ESI–FTICR–MS measurements (Figs. 5 and 6, for tables of the masses calculated and found: see Supplementary Material Tables S4–S6).
Mass spectra of a mixture of the dsDNA ON-1∙ON-2 and the seco-drug 2 in a ratio of 1:1 incubated in water at 25 °C. a 0 h of incubation, CapExit voltage −100 V; b 24 h of incubation, CapExit voltage −100 V; c 24 h of incubation, CapExit voltage −150 V; d 24 h of incubation, CID-MS/MS of ON-1–A. ON-1*: ON-1 alkylated by drug 3, ON-1-A: ON-1 after cleavage of adenine, A*: adenine alkylated by the drug 3, w 5 (ON-1) and a 10 -B 10 (ON-1): fragments of ON-1–A indicating alkylation at A-10 of ON-1
Whereas the reaction mixture after 0 h of incubation shows only the signals of the seco-drug 2, the drug 3 and of the two oligonucleotides ON-1 and ON-2 (Fig. 6a), additional peaks of the covalent adduct ON-1*, the depurinated oligonucleotide ON-1–A and the alkylated nucleobase adenin A* could be observed after 24 h of incubation (Fig. 6b).
Applying a CapExit voltage of −100 V, the alkylated oligonucleotide ON-1* underwent a partial fragmentation affording the depurinated oligonucleotide ON-1–A [13-15]. Increasing the CapExit voltage to −150 V induced a further fragmentation of the depurinated oligonucleotide ON-1–A to provide the secondary fragments w 5 (ON-1) and a 10 -B 10 (ON-1) by CSD (Fig. 6c).Footnote 1 The same fragmentation could also be achieved by CID of ON-1–A in an MS/MS experiment using argon as the collision gas (Fig. 6d, for calculated and found masses: see Supplementary Material Table S7). The fragmentation pattern clearly shows that the alkylation took place selectively at A-10 of the single-strand ON-1 of the double-stranded oligonucleotide, which is situated at the 3′ end of an AT-rich region (Fig. 7). This selectivity was also found for the natural products CC-1065 and duocarmycin SA [9-11].
Fragmentation of the alkylated oligonucleotide ON-1* into A*, w 5 (ON-1) and a 10 -B 10 (ON-1) [16]
The modification of the nucleobase weakens the N-glycosidic bond, favouring a depurination of ON-1* under participation of the neighbouring O-atom to afford the primary fragments ON-1–A and A* under mild ionisation conditions (Fig. 8). Using harsher conditions, i.e. higher capillary exit voltage, the depurinated oligonucleotide ON-1–A degrades via 1,2-elimination to give the secondary fragments w 5 (ON-1) and a 10 -B 10 (ON-1) that are indicative of the exact position of the alkylation reaction, namely the nucleobase A-10 of ON-1 (Fig. 7) [13-15].
Based on the peak intensities of the corresponding isotope peaks including all sodium adducts, the percentage of alkylation per hour (A t) was calculated considering an incubation time of up to 6 h with measurements in triplicate being performed every hour (for data, see Supplementary Material Table S8). Calculations of the alkylation rate using the deconvoluted spectra gave the same results as were obtained using the corresponding native spectra. According to Eq. 1, the alkylation rate was determined to be approximately 10% per hour (Fig. 9).
- \( \overline {Q^t} \) :
-
median ratio of the intensities of the corresponding isotope peaks and sodium adducts of ON-1 to the respective signals of ON-2 at the incubation time t
- \( {\bar Q^0} \) :
-
median ratio of the intensities of the corresponding isotope peaks and sodium adducts of ON-1 to the respective signals of ON-2 at the incubation time 0 h.
Compared to previous studies based on mass spectrometry [16-21], this procedure allows the determination of the alkylation efficiency and the alkylation position without prior purification and enrichment of the products using HPLC before the mass spectrometric investigations [12]. Furthermore, in comparison to other methods for the analysis of DNA–drug complexes, such as NMR spectroscopy [22, 23] and X-ray crystal structure analysis [24], as well as PCR stop assay, DNAseI footprinting and polyacrylamide gel electrophoresis after thermally induced strand cleavage [25-29], the procedure offers some convenient advantages like a fast determination of alkylation efficiencies and sequence selectivities with low sample amounts and without prior sample manipulation.
To confirm the results obtained by direct ESI–HRMS measurements, HPLC of the reaction mixtures followed by ESI–HRMS of the eluted fractions was performed. Again, the alkylation rate was approximately 10% per hour and the reaction took place selectively at A-10 of ON-1. When the reaction mixture was incubated for 24 h and heated up to 95 °C for 2 h prior to the HPLC separation and ESI–HRMS analyses of the eluates, cleavage of the alkylated oligonucleotide ON-1* into the fragments w 5 (ON-1) and d 9 (ON-1) occurred (Fig. 10, for tables of the masses calculated and found: see Supplementary Material Tables S9). Thus, the alkylation took place selectively at A-10 of ON-1; however, a slightly different mechanism of fragmentation of the alkylated oligonucleotide ON-1* in solution (Fig. 11) as compared to the fragmentation in the mass spectrometer (Fig. 8) was observed.
HPLC chromatogram of the reaction mixture of the dsDNA ON-1∙ON-2 and the seco-drug 2 in a ratio of 1:1 after incubation at 25 °C for 24 h and heating to 95 °C for 2 h. w 5 (ON-1) and d 9 (ON-1): characteristic fragments of ON-1 indicating alkylation at A-10 of ON-1; ON-1–A+H 2 O∙ON-2: dsDNA after cleavage of adenine in ON-1 and addition of water; ON-1∙ON-2: dsDNA; A*: adenine alkylated by the drug 3
Fragmentation of the alkylated oligonucleotide ON-1* in solution to give the depurinated oligonucleotide ON-1–A+H 2 O and the alkylated nucleobase A*. Depurination is followed by isomerisation of ON-1-A+H2O and strand cleavage at the depurinated site to afford w 5 (ON-1) and 8. Cleavage of the residues of the sugar moiety in 8 affords d 9 (ON-1) and 9
In solution, the alkylated nucleobase A* is supposed to be cleaved from the oligonucleotide under subsequent attack of water to give the hemiacetal ON-1–A+H 2 O. This hemiacetal isomerises to the corresponding aldehyde, which is prone to a 1,2-elimination of w 5 (ON-1) followed by a 1,2-elimination of d 9 (ON-1) [13-15].
Notably, the non-covalent interactions between the unmodified DNA oligonucleotides (ON-1∙ON-2) as well as of the slightly modified DNA oligonucleotides (ON-1–A+H 2 O∙ON-2) remained intact during the HPLC separation. Only after strand cleavage the retention times were notably different, indicating the dissociation of the modified double-stranded DNA into the single-strand ON-2 and the fragments w 5 (ON-1) and d 9 (ON-1) of the modified oligonucleotide ON-1.
In contrast to seco-drug 2, prodrug 1 showed only a very low reactivity against the dsDNA ON-1∙ON-2 (Fig. 12, for tables of the masses calculated and found: see Supplementary Material Tables S10 and S11). In a ratio of 1:1 (Fig. 12a), nearly no alkylation could be detected and even after 24 h of incubation of the dsDNA with prodrug 1 in a ratio of 1:5, less than 10% of the DNA had reacted to the respective covalent adduct (Fig. 12b). During the incubation with the DNA, no cleavage of the sugar moiety in 1 occurred, so that a direct nucleophilic substitution of the chlorine atom in prodrug 1 by the DNA took place. Interestingly, both DNA oligonucleotides were alkylated to give ON-1* and ON-2* as well as the depurinated oligonucleotides ON-1-A and ON-2–A, respectively.
Mass spectra of mixtures of the dsDNA ON-1∙ON-2 and prodrug 1 in a ratio of a 1:1 and b 1:5 after incubation of the respective reaction mixture for 24 h at 25 °C; CapExit voltage −100 V. ON-1*:ON-1 alkylated by prodrug 1; ON-2*:ON-2 alkylated by prodrug 1; ON-1-A: ON-1 after cleavage of adenine; ON-2-A: ON-2 after cleavage of adenine; w 5 (ON-1) and a 10 -B 10 (ON-1): fragments of ON-1–A; w 8 (ON-2) and a 7 –B 7 (ON-2): fragments of ON-2–A
Direct ESI–HRMS measurements under harsher conditions (CapExit voltage −150 V) led to an enhanced fragmentation of the depurinated oligonucleotides to provide the characteristic fragments A* (Fig. 13a), w 5 (ON-1) and a 10 –B 10 (ON-1) as well as w 8 (ON-2) and a 7 –B 7 (ON-2) (Figs. 13b and 14).
Obviously, in the small amount of alkylated dsDNA obtained by reaction of the dsDNA with an excess of prodrug 1, reaction had taken place at A-10 of the oligonucleotide ON-1 and at A-7 of the oligonucleotide ON-2 (Fig. 14). Thus, prodrug 1 seems to bind in the same binding mode and orientation to the DNA double-strand as drug 3, but, owing to the greater flexibility of the alkylating 1-chloroethyl moiety in prodrug 1 in comparison to the cyclopropyl moiety in 3, alkylation of both strands was possible (Fig. 15).
Conclusions
The results show that 1 is very suitable as prodrug for the use in ADEPT since it is not converted into the corresponding drug 3 in unmodified cell culture medium on the one hand, but on the other hand it is transformed efficiently into the seco-drug 2 in the presence of β-d-galactosidase. 2 cyclises rapidly to give the drug 3 in situ, which acts as a very potent alkylating agent of double-stranded DNA, whereas prodrug 1 shows only a very low alkylation efficiency. Since the alkylation efficiency correlates very well with the cytotoxicity in cell culture investigations, it is supposed that the drug’s high biological activity derives from the alkylation of cellular DNA.
The combination of all techniques applied led to a better understanding of the mode of action of the new compounds. This is a prerequisite for successfully studying the transformation and metabolism of 1 and its analogues in living organisms. Furthermore, a novel method for the direct investigation of the alkylation of DNA by alkylating agents using ESI–FTICR–MS was developed, which can be used to estimate relative alkylation efficiencies as well as the sites of alkylation of the DNA by different alkylating agents. The latter is based on a characteristic mode of fragmentation of the modified DNA oligonucleotides induced by CSD or by CID. In addition, mechanisms explaining the different fragmentation of alkylated oligonucleotides in the mass spectrometer and in solution are provided.
Notes
The nomenclature used for DNA fragments is based on a suggestion from S. McLuckey [15].
Abbreviations
- A:
-
Adenine
- ADEPT:
-
Antibody-directed enzyme prodrug therapy
- AuC:
-
Area under the curve
- CID:
-
collision-induced dissociation
- CSD:
-
capillary-skimmer dissociation
- DNA:
-
Deoxyribonucleic acid
- ESI–FTICR–MS:
-
electrospray–Fourier transform ion cyclotron resonance–mass spectrometry
- HPLC–MS:
-
high-pressure/performance liquid chromatography–mass spectrometry
References
Tietze LF, Krewer B (2009) Anti-Cancer Agents Med Chem 9:304–325
Tietze LF, Feuerstein T (2003) Curr Pharm Des 9:2155–2175
Bagshawe KD (1987) Br J Cancer 56:531–532
Bagshawe KD (2006) Expert Rev Anticancer Ther 6:1421–1431
Tietze LF, Major F, Schuberth I, Spiegl DA, Krewer B, Maksimenka K, Bringmann G, Magull J (2007) Chem Eur J 13:4396–4409
Tietze LF, Major F, Schuberth I (2006) Angew Chem Int Ed 45:6574–6577
Martin DG, Biles C, Gerpheide SA, Hanka LJ, Krueger WC, McGovren JP, Mizsak SA, Neil GL, Stewart JC, Visser J (1981) J Antibiot 34:1119–1125
Ichimura M, Ogawa T, Takahashi K, Kobayashi E, Kawamoto I, Yasuzawa T, Takahashi I, Nakano H (1990) J Antibiot 43:1037–1038
Boger DL, Johnson DS (1996) Angew Chem Int Ed 35:1438–1474
Hurley LH, Needham-VanDevanter DR (1986) Acc Chem Res 19:230–237
Hurley LH, Reynolds VS, Swenson DH, Petzold GL, Scahill TA (1984) Science 226:843–844
Tietze LF, Krewer B, Frauendorf H, Major F, Schuberth I (2006) Angew Chem Int Ed 45:6570–6574
Hofstadler SA, Sannes-Lowery KA, Hannis JC (2005) Mass Spectrom Rev 24:265–285
Banoub JH, Newton RP, Esmans E, Ewing DF, Mackenzie G (2005) Chem Rev 105:1869–1915
McLuckey S, Habibi-Goudarzi S (1993) J Am Chem Soc 115:12085–12095
Fitzner A, Frauendorf H, Laatsch H, Diederichsen U (2008) Anal Bioanal Chem 390:1139–1147
Colgrave ML, Iannitto-Tito P, Wickham G, Sheil MM (2003) Aust J Chem 56:401–413
Beck JL, Colgrave ML, Ralph S, Darzi S (1993) J Am Chem Soc 115:12085–12095
Beck JL, Colgrave ML, Ralph SF, Sheil MM (2001) Mass Spectrom Rev 20:61–87
Iannitti-Tito P, Weimann A, Wickham G, Sheil MM (2000) Analyst 125:627–634
Iannitti P, Sheil MM, Wickham G (1997) J Am Chem Soc 119:1490–1491
Smith JA, Bifulco A, Case DA, Boger DL, Gomez-Paloma W, Chazin J (2000) J Mol Biol 300:1195–1204
Eis PS, Smith JA, Rydzewski JM, Bifulco A, Case DA, Boger DL, Chazin J (1997) J Mol Biol 272:237–252
Kennard O, Hunter WN (1991) Angew Chem Int Ed Engl 30:1254–1277
Bando T, Narita A, Sasaki S, Sugiyama H (2005) J Am Chem Soc 127:13890–13895
Kupchinsky S, Centioni S, Howard T, Trzupek J, Roller S, Carnahan V, Townes H, Purnell B, Price C, Handl H, Summerville K, Johnson K, Toth J, Hudson S, Kiakos K, Hartley JA, Lee M (2004) Bioorg Med Chem 12:6221–6236
Boger DL, Schmitt HW, Fink BE, Hedrick MP (2001) J Org Chem 66:6654–6661
Zhang Z, Poirier MC (1997) Chem Res Toxicol 10:971–977
Pommier Y, Kohlhagen G, Bailly C, Waring M, Mazumder A, Kohn KW (1996) Biochemistry 35:13303–13309
Acknowledgements
This research was supported by the Deutsche Forschungsgemeinschaft and the Fonds der Chemischen Industrie. B.K. thanks the Deutsche Telekom Foundation for a Ph.D. scholarship. The authors thank F. Major for providing 1 and 2 and I. Schuberth for determining the IC50 values of 1 and 2.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
Awarded a poster prize at the 27th International Symposium on Chromatography, 21–25th September 2008 in Münster, Germany
Electronic Supplementary Materials
Below is the link to the electronic supplementary material.
ESM 1
(PDF 109 kb)
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Tietze, L.F., Krewer, B. & Frauendorf, H. Investigation of the transformations of a novel anti-cancer agent combining HPLC, HPLC–MS and direct ESI–HRMS analyses. Anal Bioanal Chem 395, 437–448 (2009). https://doi.org/10.1007/s00216-009-2963-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-009-2963-x