Abstract
The analysis of mutations that are associated with the occurrence of drug resistance is important for monitoring the antiretroviral therapy of patients infected with human immunodeficiency virus (HIV). Here, we describe the establishment and successful application of Arrayed Primer Extension (APEX) for genotypic resistance testing in HIV as a rapid and economical alternative to standard sequencing. The assay is based on an array of oligonucleotide primers that are immobilised via their 5′-ends. Upon hybridisation of template DNA, a primer extension reaction is performed in the presence of the four dideoxynucleotides, each labelled with a distinct fluorophore. The inserted label immediately indicates the sequence at the respective position. Any mutation changes the colour pattern. We designed a microarray for the analysis of 26 and 33 codons in the HIV protease and reverse transcriptase, respectively, which are of special interest with respect to drug resistance. The enormous genome variability of HIV represents a big challenge for genotypic resistance tests, which include a hybridisation step, both in terms of specificity and probe numbers. The use of degenerated oligonucleotides resulted in a significant reduction in the number of primers needed. For validation, DNA of 94 and 48 patients that exhibited resistance to inhibitors of HIV protease and reverse transcriptase, respectively, were analysed. The validation included HIV subtype B, prevalent in industrialised countries, as well as non-subtype B samples that are more common elsewhere.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
AIDS—acquired immunodeficiency syndrome—was first reported in 1981 and has since become a worldwide pandemic. Currently some 40 million people are HIV-infected (http://www.who.int/). Antiviral treatment is based on the use of drugs which inhibit HIV reverse transcriptase (RT) and protease (PR). The emergence of resistant viruses is a major obstacle to the long-term effective and successful therapy of HIV-infected patients. Antiviral drug resistance is caused by the presence of mutations in the PR and RT coding regions of the virus that reduce drug susceptibility compared to wild-type viruses (see e.g. [1]). Drug resistance can be determined by genotypic tests that identify resistance-related mutations in the viral genome [2]. Therefore, genotypic resistance testing has become an increasingly important process when monitoring the antiretroviral therapy of patients infected with HIV [3–7].
HIV genotypic resistance data are mostly obtained during the clinical routine by dideoxy sequencing [8] which is a time-consuming process. Alternative methods for genotypic resistance testing are needed. Therefore, several different approaches have been described in the last few years, based on hybridisation [9], primer extension [10] or ligation methods [11]. Because of the tremendous variability of the viral genome, the two last approaches were restricted to a few selected mutations, however. In consequence, primary mutations were assayed but secondary mutations were not included. Primary mutations are directly responsible for resistance. Secondary or accessory mutations often cause resistance when present in combination with other mutations or compensate for the diminished replicate activity that can be associated with drug resistance [2]. The assay reported by Gonzalez et al. [9] covered a wide range of mutations based on the microarray technology developed by Affymetrix. Even with the large number of oligonucleotides that can be placed on this chip, however, it is difficult to achieve clinically reliable results, since the analysis is based merely on hybridisation discrimination and is therefore prone to being obscured by the high degree of sequence variance in HIV.
Here, we show the development and validation of a cost-effective and rapid HIV resistance test that allows the detection of the mutations in PR and RT that are reported as being resistance-related by the Stanford HIV database http://hivdb.stanford.edu/). It is based on the Arrayed Primer Extension method (APEX; [12]), a process used to detect single nucleotide polymorphisms in a target DNA. By virtue of the combination of the discriminative effect of the hybridisation process and the base-pairing specificity of the DNA-polymerase used for the extension reaction, the overall accuracy of the reaction can be increased 10- to 100-fold compared to merely hybridisation-based assays [13, 14]. Template-dependent extension of the arrayed primers in the presence of the four different dideoxynucleotides, each labelled with a particular fluorophore, yields sequence information. Mutations can be detected by a change in the signal colour at the respective array position. In order to minimise the number of oligonucleotides while at the same time being able to analyse all relevant mutations in PR and RT, the applicability of degenerated oligonucleotides was also studied. A primer is called degenerated if several bases are possible at some of its sequence positions. Degenerated oligonucleotides have been found to be useful in PCR [15] and hybridisation assays (e.g. [16]), for example. The final objective was an adaptation of the APEX technology to the challenges provided by the high mutability of the HIV genome and the development of a quick, efficient and inexpensive genotypic test system for high-throughput clinical applications.
Materials and methods
Sample collection
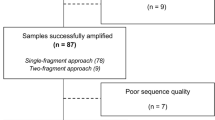
To evaluate the APEX-based genotypic resistance assay, 94 and 48 HIV-1 positive clinical samples from patients that exhibited resistance to inhibitors of HIV protease and reverse transcriptase, respectively, were analysed. The patient-derived PCR-products were chosen randomly from routine clinical testing at the Department of Virology, University of Heidelberg. For all PCR products, the sequences were determined by Sanger sequencing using the ViroSeq genotyping kit (Abbott, Wiesbaden, Germany). Phylogenetic analysis showed that 76% (71/94) of the PR samples and 87% (41/48) samples of the RT region belonged to HIV-1 subtype B. The non-subtype B samples consisted of A, C, F1, CRF01_AE, CRF01_AG, CRF02_AG and CRF06_cpx (Rega HIV-1 Subtyping Tool, [17]).
Template preparation
For re-amplification, 5-μl aliquots of the original PCR-products were used as template. The 50 μl reaction mix consisted of 0.25 mM dNTPs, 0.3 mM each of forward and reverse primer, 1.25 U Thermoprime Plus DNA polymerase and reaction buffer (ABgene, Hamburg, Germany). 15% of the dTTP was replaced with dUTP in order to introduce dUTP randomly. For amplification, primers d(GGGAAGATCTGGCCTTCCTACAA) and d(GGGCCATCCATTCGTGGC) were used for PR and d(CACCTGTCAACATAATTGGA) and d(ACTGTCCATTTATCAGGATG) for the RT region. The PCR primers were synthesised by biomers (Ulm, Germany).
PCR began with an initial denaturation step at 94 °C for 2 min followed by 30 cycles at 94 °C for 30 s, 56 °C for 30 s and 72 °C for 2.5 min, and a final extension at 72 °C for 10 min. PTC-200 ThermoCyclers (MJ Research, Waltham MA, USA) were used for all amplifications. The expected fragment lengths of 559 bp (PR) and 780 bp (RT) were checked on 2% agarose gels. One hundred microlitres of PCR mix were concentrated and purified with a QIAquick PCR Purification Kit (Qiagen, Hilden, Germany), as recommended by the manufacturer, and eluted in 30 μl H2O.
Finally, 20 μl of each PCR product was fragmented by incubation with 0.1 U uracil-N-glycosylase in a reaction buffer of 50 mM Tris/HCl pH 9, 20 mM (NH4)2SO4) at 37 °C for 1 h. The enzyme was deactivated at 95 °C for 10 min, and an aliquot of digested DNA was again checked on an agrose gel.
Production of the oligonucleotide microarrays
All oligonucleotides were purchased from biomers (Ulm, Germany) or MWG (Ebersberg, Germany). Each one was 50 nucleotides in length, containing a C6-aminolinker and a 20-25mer oligo-dT spacer at the 5′-end. Attachment to epoxysilane-coated glass slides (Schott Nexterion Slide E, Mainz, Germany) occurred via the 5′-C6-aminolinker to ensure the accessibility of the 3′-terminus for the enzymatic reaction. The final spotting solution contained 50 μM oligonucleotide in 0.05 M sodium carbonate, 0.1 M sodium citrate, pH 9.5, with 3 M betaine.
In addition to the HIV-specific primers, two kinds of controls were included in the chip layout. First, 25-mer oligonucleotides that carried at their 3′-end the fluorescence labels Cy3, Cy5, TexasRed or Fluorescein, respectively, were utilised as positional controls (5′-TGGGTATAAACGATTTCGTAGATAA-dye). A mixture of the four differently labelled oligonucleotides (12.5 μM each) was spotted onto the microarray. Second, to control the primer extension reaction an oligonucleotide was applied that forms a hairpin structure (NACGACCGACTTTAGGTCCCTCAATTTTTGAGGGACCTAAAGTCGGTCGT), thus acting as both primer and template. In analogy to the molecules that represent HIV sequences, all controls had a C6 aminolinker and an oligo-dT spacer at their 5′-ends.
For microarray production, the SDDC-2 Chip Writer contact printing robot (Engineering Services, Toronto, ON, Canada) was used. Equipped with SMP 3-pins (TeleChem, Sunnyvale, CA, USA), each contact delivered a volume of approximately 1 nl. All oligonucleotides were spotted in duplicate. After spotting, slides were incubated at 60 °C for 1 h and stored at room temperature for at least 24 h.
Arrayed primer extension
Directly before an APEX reaction was performed, the microarrays were washed twice in H2O at 95 °C to remove residual spotting solution. To block the surface, slides were incubated in 100 mM NaOH for 10 min at room temperature and washed again in H2O at 95 °C. The primer extension was carried out directly on the slide surface in a reaction chamber formed by a 20 μl frame-seal (ABgene, Hamburg, Germany).
As estimated from a comparison to a 100 bp DNA marker ladder (MBI Fermentas, St. Leon-Roth, Germany), 600–800 ng of PCR product were used in each APEX reaction. The 20-μl reaction mix contained 10 μl of fragmented PCR product, 4 U ThermoSequenase DNA polymerase (Amersham, Freiburg, Germany), ThermoSequenase reaction buffer (Amersham) and 1.25 μM each of ddATP-TexasRed, ddCTP-Cy3, ddGTP-Fluorescein and ddUTP-Cy5 (Perkin Elmer, Boston, MA, USA). The mix of buffer, labelled ddNTPs and target DNA were first denatured for 10 min at 95 °C before adding the DNA polymerase. The mix was subsequently loaded onto the microarray, and the reaction chamber was covered with a plastic film.
The slides were placed into a ThermoCycler PTC-200, TwinTower (MJ Research, Waltham, MA, USA). Temperature profiles were used as shown in Table 1. After the primer extension reaction, the slides were immediately washed in water at 95 °C, for 3 min in 0.3% Alconox (Alconox, White Plains, NY, USA), again in water at 95 °C, and finally in 30 mM NaCl, 3 mM Na3 citrate (pH 8.5) in order to obtain optimal pH conditions for all four fluorescent dyes. Slides were dried in an air flow.
Signal analysis
Signal detection was performed by confocal four-colour laser scanners (ScanArray 4000XL and ScanArray 5000, Perkin Elmer, Boston, MA, USA). Intensities were measured using GenePix 4.0 quantification software (Axon Instruments, Union City, CA, USA). The data were normalised based on the quantum yields of the four fluorescent dyes and different hybridisation properties of the oligonucleotides using a software tool developed in-house. The output files presented a base call for each nucleotide position and allowed a direct comparison with the reference sequences obtained by Sanger sequencing.
Results and discussion
Optimisation of APEX conditions
The application of APEX for HIV resistance testing required some modifications to the reaction conditions as described by Kurg et al. [12] for standard genotyping. Each oligoucleotide had a 20–25mer portion of oligo-d(T) at its 5′-end to reduce steric hindrance effects. Spotting the oligonucleotides in sodium carbonate buffer (pH 9) produced the best signal-to-background ratios. The addition of 3 M betaine to the spotting solution improved spot homogeneity [18]. Furthermore, in contrast to the temperature profile described by Kurg et al. [12], primer extension was not carried out as a one-step reaction at 48 °C. At least three steps were needed for denaturation, hybridisation and specific primer extension in order to obtain significant signal intensities at the PR oligonucleotides (Table 1). Dissociation temperatures were even more variable for the RT oligonucleotide primers. Therefore a multi-step gradient profile was established (Table 1).
Degenerated oligonucleotide primers
Because of its replication cycle, HIV shows immense sequence heterogeneity [19–22]. To effectively analyse a wide range of samples, it is necessary to represent this very high sequence variety on the microarray. In order to reduce the number of oligonucleotides needed, the effects of wobble positions (a mixture of bases at the particular nucleotide) and substitution with inosine was investigated. The most polymorphic region of the PR gene was used as a model. It is located around codon 71. In total, 13 positions of the 20-mer primer that was used to query the nucleotide at sequence position 211 are not conserved. A probe representing the wildtype sequence was used as a control. In derivative molecules, more and more variable positions were replaced by degenerate bases, starting either from the 5′-end or 3′-end (Table 2). In one oligonucleotide (W13), all variable positions were degenerated.
Oligonucleotide extension was performed at the conditions described in the “Materials and methods” section. Using a wild-type DNA-template, Cy3-ddCTP was incorporated. As shown in Fig. 1, increasing the number of degenerated positions within the primer sequence led to a decrease in the signal. Oligonucleotides with 1–5 wobble positions at the 5′-terminus exhibited reduced but still significant signal intensities compared to the unmodified probe (Fig. 1a). In contrast, the introduction of even one wobbling base at the 3′-end (3W1) reduced signal intensity dramatically. No significant signal could be detected with oligonucleotides that had more than one degenerated position near the 3′-end (Fig. 1b). Additional experiments with inosine and other sequences produced similar results (data not shown). Even though inosine should base-pair with all four bases, although to a reduced degree, its introduction close to the 3′-end resulted in a complete loss of signal.
Impact of degenerated primers on APEX. Wobble sequences were introduced into the wild-type sequence at 13 positions of variable bases of a 20-mer primer starting from the 5′-end (a) or the 3′-end (b), as shown in Table 2. The effect on signal intensities that resulted from a primer extension reaction on wild-type DNA is presented
Based on these results, the oligonucleotides for the microarray were designed in such a way that no more than four wobble nucleotides or inosines were introduced as close to the 5′-terminus as possible. This strategy allowed an ~16-fold reduction in the number of primers needed on the chip for genotypic testing of all major resistance sites in HIV.
Chip design
The final assay was designed to detect the key mutations which are associated with the development of drug resistances in HIV-1. The selection of codons within the HIV-1 PR and RT coding regions was based on the drug resistance summaries in the Stanford HIV database http://hivdb.stanford.edu/), which had 15,380 entries. In essence, there are 26 relevant codons (78 nucleotide positions) in the protease gene and 33 codons (99 nucleotide positions) in the reverse transcriptase gene. The design of the oligonucleotides was based on the Los Alamos HIV sequence database http://hiv-web.lanl.gov/content/index). All sequence variations described as resistance-related and present in more than 5% or 10% of the cases of resistance to inhibitors of PR or RT, respectively, were covered by oligonucleotides on the chip. In total 368 PR-and 523 RT-specific oligonucleotides were spotted in duplicate onto the microarray (for sequences see the “Electronic Supplementary Information”). Fluorescently labelled oligonucleotides were used for positional control. Additionally, oligonucleotides that are able to self-extend by forming hairpin structures were introduced into the chip layout as a template-independent control for the primer extension reaction; they represent template and primer at the same time (for sequences see “Materials and methods”). Figure 2 shows typical results from an APEX reaction on the PR oligonucleotides. The microarray was scanned at the wavelengths correlating to the four fluorescence dyes attached to the four dideoxynucleotides.
Initial validation of the APEX assay in comparison to standard sequencing
In the analysis, patient samples were investigated that had been obtained from ongoing routine testing at the Department of Virology of the University of Heidelberg. The analysis was approved by the local ethics committees at the University of Heidelberg, and informed consent was obtained from all patients. The set used for validation was randomly chosen and independent of subtype, drug regimens or the presence of special genotypes. A total of 7,332 relevant nucleotide positions were analysed by APEX for the HIV PR region (78 nucleotides × 94 patient samples) and 4,752 for the HIV RT gene (99 nucleotides × 48 patient samples) and compared with the results obtained by standard Sanger sequencing as a reference method.
For the PR gene, 92% (6,750 nucleotides) of the sequence information was in full agreement, while 6% (443) did not match entirely. About 2.5% (165) of the conflicting positions were due to unassigned nucleotides. For codons 10 and 36, for example, detection efficiency was low. Weak signal intensities were defined as (false) negatives, since it was impossible to determine the sequence with the stringent filter criteria applied. Another 2% (139) of the positions partially matched in APEX and sequencing since they yielded more than one base. Nucleotide mixtures that are only detected in the APEX assay but not in standard sequencing could occur because of the higher sensitivity of the APEX method to the presence of two bases at one site. The analysis software for standard sequencing often suppresses the weaker signal of the two. Different results between APEX and Sanger sequencing were definitely obtained, mostly for secondary mutations, especially codons 63 and 71 in the protease. These positions are located within the most polymorphic region; more specific rather than degenerated primers are needed at these sites. Concordance for the investigated RT samples was 86% (4,099), partial concordance 4% (161) and discordance 10% (492). A portion of 5% (234) of the positions identified as different were false negatives. Figure 3 shows the degree of concordance between APEX and Sanger sequencing for the individual 26 codons in the PR gene and 33 codons of the RT gene.
Concordance of the APEX results with data from standard sequencing. DNA from 94 and 48 patients that exhibited resistance to inhibitors of HIV protease and reverse transcriptase were analysed in 26 resistance-related codons of the PR and 33 codons of the RT genes. The data were compared to results obtained by standard Sanger sequencing of the same samples
Phylogenetic analysis of RT and PR sequences
At least nine different genetic HIV-1 subtypes and several recombinant forms exist [23]. Since subtype B is the predominant subtype in highly industrialised countries, most HIV-1 sequence data in databases is subtype B, even though subtype B is responsible for only 12% of global infections [24]. Since sequences from the Los Alamos HIV database were used as basis for the chip design, the microarray currently represents mainly subtype B sequences.
Validation of our APEX genotyping assay was done with subtype B as well as non-subtype B samples. A phylogenetic analysis according to De Oliveira et al. [17] of the PR and RT samples used in our chip validation process showed that 71 of 94 (PR) and 41 of 48 (RT) sequences belonged to subtype B. The others consisted of subtypes A, C, F1 and recombinant A/G and A/E viruses. Table 3 shows a comparison of the results for subtype B and non-subtype B samples. Focusing only on subtype B samples, concordance values of 94% and 88% were obtained for PR and RT, respectively. In comparison, the values for non-subtype B samples were 88% and 84%. Although the concordance of the APEX results with the reference method was a little lower for non-subtype B samples, the oligonucleotide set selected purely on subtype B sequences produced a base calling of sufficient accuracy to be used for non-subtype B samples.
Improvement in APEX detection
In general, the quality of the results from an APEX reaction depends on the functioning of the relevant primers. It is well documented that particular sequences may not perform well for various reasons. A promising and simple strategy for solving APEX detection problems is a modification of the microarray design by replacement and rejection of the ill-performing oligonucleotides. The analysis reported above was based on an analysis of one DNA strand only. Replacement of a primer by another oligonucleotide that queries the corresponding position on the second DNA strand is bound to improve overall detection efficiency, since an entirely different primer sequence is used, which in most cases is likely to perform well. This was demonstrated for the analysis of the PR gene. For ten poorly detected positions in codons 23, 30, 47, 60, 62, 63, 76 and 93, oligonucleotides were replaced with primers for the second DNA strand. Four samples with below-average concordance values (less than 92%) were tested again in eight repetitions on this second set of oligonucleotides. For all samples, the new base calling accuracy at the individual sites was in agreement with the sequence data. Although new primers had been designed for only ten positions, even the overall base calling accuracy increased significantly compared to the unmodified primer set (Fig. 4). While the replacement of individual primer oligonucleotides is time-consuming using the a microarray system based on spotting as reported here, the flexibility of micromirror-controlled in situ synthesis could be utilised for this purpose [25]. In this way, an optimisation process involving an iterative cycle of probe design and experimental evaluation can be performed rather quickly and cost-efficiently.
Improving concordance by replacing ill-performing primers. For ten positions in codons 23, 30, 47, 60, 62, 63, 76 and 93 that were not detected well with the initial primer set, oligonucleotides were designed to query the same sites on the second DNA strand. Four patient samples were analysed eight times. Comparison of the overall performance of the initial primer set (blue) with the modified primer set (red) indicates a significant improvement, even though only a few primers had been replaced
Data analysis
The information gained from the experiments was subjected to a correspondence cluster analysis [26], which is an explorative computational method for the investigation of associations between variables, such as polymorphisms, and the individual samples, in a multidimensional space. It simultaneously displays data for two (or more) variables in a low-dimensional projection, thus revealing associations between them. Figure 5 shows the results of clustering the data from the PR analysis. The black dots represent individual polymorphisms, the red squares the individual patient samples. Co-localisation of samples in the blot is indicative of a strong association. It is apparent that the samples separate into two subgroups. Polymorphisms associated to one of these groups are plotted in the same direction. There are only a few polymorphisms which do not contribute to the discrimination of the samples, located at the center of the plot (point 0/0) or along a line that extends above or below this point. Data from more patients with good clinical annotation is required in order to identify in a statistically meaningful manner the variable that causes clear division into (at least) two subgroups.
Conclusions
This study demonstrates the successful application of APEX to genotypic drug resistance testing in HIV. Because of the use of degenerated primers, the number of sensor molecules could be kept relatively small. Nevertheless, the assay is not restricted to the detection of primary mutations in HIV. Instead, all resistance-related mutations presently known to be associated with drug resistance could be studied in a single experiment. With a slight decrease in the number of degenerated bases introduced into primers that bind to highly polymorphic sites, and by using primers for both strands, a specificity and accuracy similar to an analysis by standard Sanger sequencing can be achieved. Considering that polymorphisms in the HIV PR and RT genes are common, even among therapy-naïve patients, it could happen in rare cases that an individual primer on the microarray may not work for an individual patient—even with an optimised set of oligonucleotide primers—because of the presence of a mutation that is particular to this patient. However, given the fact that there seem to be subgroups of polymorphisms that are highly associated with each other, such an event would most likely not influence the overall outcome of the analysis. Even with the current setup, both HIV subtype B and non-subtype B samples can be analysed. Furthermore, the flexible array design allows an uncomplicated inclusion of additional oligonucleotides if necessary for this or other special applications. Another attractive aspect of the assay is that the entire procedure takes only a few hours, including data analysis. Therefore, this microarray approach allows genotypic resistance testing in a high-throughput manner with an accuracy that seems sufficient for routine clinical application.
References
D’Aquila RT (1995) Clin Diagn Virol 3:299–316
Shafer RW (2002) Clin Microbiol Rev 15:247–277
Hirsch MS, Conway B, D’Aquila RT, Johnson VA, Brun-Vezinet F, Clotet B, Demeter LM, Hammer SM, Jacobsen DM, Kuritzkes DR, Loveday C, Mellors JW, Vella S, Richman DD (1998) JAMA 279:1984–1991
Durant J, Clevenbergh P, Halfon P, Delgiudice P, Porsin S, Simonet P, Montagne N, Boucher CA, Schapiro JM, Dellamonica P (1999) Lancet 353:2195–2199
Weinstein MC, Goldie SJ, Losina E, Cohen CJ, Baxter JD, Zhang H, Kimmel AD, Freedberg KA (2001) Ann Intern Med 134:440–450
Wegner SA, Wallace MR, Aronson NE, Tasker SA, Blazes DL, Tamminga C, Fraser S, Dolan MJ, Stephan KT, Michael NL, Jagodzinski LL, Vahey MT, Gilcrest JL, Tracy L, Milazzo MJ, Murphy DJ, McKenna P, Hertogs K, Rinehart A, Larder B, Birx DL (2004) Clin Infect Dis 38:723–730
Zolopa AR (2006) J Infect Dis 194(Suppl 1):S59–S64
Sanger F, Nicklen S, Coulson AR (1992) Biotechnology 24:104–108
Gonzalez R, Masquelier B, Fleury H, Lacroix B, Troesch A, Vernet G, Telles JN (2004) J Clin Microbiol 42:2907–2912
Sugiura W, Shimada K, Matsuda M, Chiba T, Myint L, Okano A, Yamada K (2003) J Clin Microbiol 41:4971–4979
Beck IA, Mahalanabis M, Pepper G, Wright A, Hamilton S, Langston E, Frenkel LM (2002) J Clin Microbiol 40:1413–1419
Kurg A, Tonisson N, Georgiou I, Shumaker J, Tollett J, Metspalu A (2000) Genet Test 4:1–7
Syvanen AC (1999) Hum Mutation 13:1–10
Tõnisson N, Zernant J, Kurg A, Pavel H, Slavin G, Roomere H, Meiel A, Hainaut P, Metspalu A (2002) Proc Natl Acad Sci USA 99:5503–5508
Kilpatrick DR, Nottay B, Yang CF, Yang SJ, Mulders MN, Holloway BP, Pallansch MA, Kew OM (1996) J Clin Microbiol 34:2990–2996
Chizhikov V, Wagner M, Ivshina A, Hoshino Y, Kapikian AZ, Chumakov K (2002) J Clin Microbiol 40:2398–2407
DeOliveira CF, Diaz RS, Machado DM, Sullivan MT, Finlayson T, Gwinn M, Lackritz EM, Williams AE, Kessler D, Operskalski EA, Mosley JW, Busch MP (2000) Transfusion 40:1399–1406
Diehl F, Grahlmann S, Beier M, Hoheisel JD (2001) Nucleic Acids Res 29:E38
Moutouh L, Corbeil J, Richman DD (1996) Proc Natl Acad Sci USA 93:6106–6111
Preston BD, Poiesz BJ, Loeb LA (1988) Science 242:1168–1171
Roberts JD, Bebenek K, Kunkel TA (1988) Science 242:1171–1173
Perelson AS, Neumann AU, Markowitz M, Leonard JM, Ho DD (1996) Science 271:1582–1586
McCutchan FE (2006) J Med Virol 78(Suppl 1):S7–S12
Kantor R, Katzenstein D (2004) J Clin Virol 29:152–159
Pullat J, Fleischer R, Becker N, Beier M, Metspalu A, Hoheisel JD (2007) BMC Genomics 8:282
Fellenberg K, Hauser NC, Brors B, Neutzner A, Hoheisel JD, Vingron M (2001) Proc Natl Acad Sci USA 98:10781–10786
Acknowledgements
We thank Petra Elbert and Edith Daum of the diagnostics unit of the Department of Virology for sequences and PCR-products. We are grateful to Andres Metspalu at the University of Tartu and the company Asper, also located in Tartu, Estonia, for sharing information about APEX. This work was supported in part by the MolTools project consortium funded by the European Commission and a grant of the Landesstiftung Baden-Württemberg.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 43.5 kb)
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Schanné, M., Bodem, J., Gerhold-Ay, A. et al. Genotypic resistance testing in HIV by arrayed primer extension. Anal Bioanal Chem 391, 1661–1669 (2008). https://doi.org/10.1007/s00216-007-1775-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-007-1775-0