Abstract
The computational study of drug/protein interactions is fundamental to understand the mode of action of drugs and design new ones. In this study, we have developed a python code aimed at characterizing the nature of drug/amino acids interactions in an accurate and automatic way. Specifically, the code is interfaced with different software packages to compute the interaction energy quantum mechanically, and obtain its different contributions, namely, Pauli repulsion, electrostatic and polarisation terms, by an energy decomposition analysis based on one-electron and two-electron deformation densities. The code was tested by investigating the nature of the interaction between the glycine amino acid and 250 drugs. An energy-structure relationship analysis reveals that the strength of the electrostatic and polarisation contributions is related with the presence of small and large size heteroatoms, respectively, in the structure of the drug.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Proteins are implied in many biological processes in our organisms by interacting with other molecules, including drugs [1]. Pharmacology aims at getting insight into the metabolic route of the drugs once they have entered our bodies. Specifically, the drug/protein interactions have been intensively investigated in many research studies since the drug/protein binding process determines, in a great extent, the distribution, toxicity and activity of the drugs and, therefore, their therapeutic efficiency [2]. For example, the binding of different drugs to blood proteins, as albumin, controls the osmotic pressure, among other functions [3]. Hence, the study of drug/protein interactions is crucial to understand the mode of action of the drugs and to develop novel therapeutic agents with enhanced efficacy. The application of experimental techniques often remains the most trustworthy approach, but the experimental characterization of a large number of drug/protein pairs is very time-consuming and costly due to sample volume and instrumentation [4, 5]. Thus, computational methods have gained popularity since they can be applied in a systematic and efficient way and can provide mechanistic information which is not attainable by experimental measurements [4].
The complexity of biological systems and processes, such as the drug/protein binding, requires the use of approximations in the theoretical models to handle a large number of atoms and run longer simulation times, which are often required due to the slowness of the physical and chemical processes when they occur in biological media. Most of the theoretical approaches simplify the calculation of the interatomic interactions, which is the most time-consuming step in computational modelling. This is the case of classical molecular dynamics, which employs simple analytical functions, called force fields, to compute very efficiently the interactions that govern the behaviour of large systems. In the last decades, classical molecular dynamics has been employed in the theoretical investigation of a wide range of biological processes, including protein folding [6,7,8,9], drug/protein binding [10,11,12,13], diffusion of small molecules across lipid bilayers [14,15,16,17,18,19], DNA damage [20,21,22,23,24], ion conduction through ion channels [25,26,27,28,29,30], and many others.
The conventional force fields, including both fixed-charged and polarisable ones, have the advantage of introducing physical meaning into the potential energy function employed to compute the interactions. Thus, the most employed force fields, such as Amber [31], CHARMM [32], and OPLS [33], split the non-bonded interactions into electrostatic and non-electrostatic contributions, which are described by the Coulomb and Lennard–Jones potentials. The latter one has repulsive and attractive contributions, which intent to account for the Pauli repulsion and van der Waals interactions, respectively. However, conventional force fields present a clear limitation: the low flexibility of their analytical functions does not provide an accurate description of many systems and processes, especially those that were not employed in their parameterization. Therefore, the accuracy and transferability of conventional force fields are limited [34]. To circumvent these problems, machine learning force fields have been developed in the last years [35,36,37,38,39]. In this approach, a very flexible functional form able to describe any type of non-linear behavior is fitted to high-level electronic-structure energies (and sometimes also forces), which are obtained from quantum mechanical calculations. Therefore, the ability to describe the correct shape of the potential-energy surface does not rely on a predefined function with physical meaning, but on an unbiased function containing a large set of parameters that are adjusted to reproduce large electronic-structure data sets. This unbiased function is often formed by a combination of artificial neural networks organized in different layers. However, despite the convenient feature of being very flexible, the lack of physical meaning behind the force fields based on machine learning methods precludes the characterization of the interactions that control the process under investigation.
One possible way of introducing physical meaning into the machine learning model is to train the algorithm not only with interaction energies but also with their different contributions, which can be obtained from an energy decomposition analysis (EDA) scheme. For example, a neural network force field has recently been trained with interaction energy contributions from symmetry-adapted perturbation theory (SAPT) calculations with the very specific goal of reproducing intermolecular interaction energies for hydrogen-bonded complexes [40]. However, the use of such an approach requires the automatization of both, the computation of the interaction energies and decomposition into different energy contributions for a very large set of complexes, with different orientations and distances between the molecules. In this contribution, we have developed an algorithm in python with the aim of automatizing the interaction energy calculations and subsequent EDAs based on the decomposition of the one-electron and two-electron densities [41, 42], which is less computationally demanding than SAPT calculations. Specifically, the developed algorithm was applied to characterize the nature of the interactions between the glycine amino acid and a set of 250 drugs taken from the ZINC database [43]. In addition, a detailed analysis of the relation between the interaction energy contributions and the complex structure is performed.
2 Methodology and computational details
In this study, we have developed a python code aimed at automatizing the procedure of computing the interaction energy by quantum mechanical calculations followed by the analysis of the different interaction energy contributions for a large set of complexes. Specifically, the developed code is applied to the investigation of 250 complexes formed by the glycine amino acid, whose geometry has been taken from Ropo et al. [44], and 250 drugs selected from the ZINC database [43]. Since the main goal of the present study is the development and assessment of the code, and not the analysis of a large set of molecules, as initial approach, only the neutral form of the drugs and glycine was considered in the calculations. However, a more complete molecular set should include different protonation states for both the drugs and the amino acids since they will likely present different interaction energies and energy components. The general workflow of the procedure driven by the code, represented in Fig. 1, consists of different steps: (i) choice of the initial intermolecular distance between the two molecules of the complex; (ii) determination of the orientation between the molecules of the complex for which the interaction is the most favourable one; (iii) calculation of the potential energy curve for the most favourable orientation at a low level of theory; (iv) high-level energy calculations for five geometries selected around the low-level energy minimum; and (v) EDA employing the high-level electron densities for those five geometries. A detailed explanation of these steps will be given for an specific complex formed by the glycine amino acid and the 3-(2-methoxyphenoxy)propane-1,2-diol drug (see Fig. 2A). From now on, they will also be called molecule 1 and molecule 2, respectively, for simplicity.
In order to avoid a very large repulsive or a very low attractive interaction between the molecules of the complex, an appropriate initial intermolecular distance between the molecules has to be chosen in step (i). This distance is defined between the geometric centres of the two molecules, and it is calculated in the following manner. First, the interatomic distances between all the atoms within each of the molecules is calculated. Then, the largest interatomic distance for each molecule is selected (\(d_{1}\) and \(d_{2}\)). These two distances are then employed as the diameters of two generated spheres centred on the geometric centres of the molecules (see Fig. 2A). In order to avoid overlaps between atoms of different molecules, each sphere is enlarged by adding to its radius twice the van der Waals radius of the largest atom of the corresponding molecule. In the case of the example complex shown in Fig. 2A, for molecule 1 the largest distance (or diameter d1) corresponds to the distance between the oxygen atom forming the C=O double bond of the carboxyl group and the hydrogen atom of the amino group, which is 4.33 Å. Then, twice the van der Waals radius of the carbon atom (1.70 Å), which is the largest atom of the molecule, is added to sphere radius. In the case of molecule 2, the maximum distance is found between two hydrogen atoms (see Fig. 2A), and the atom with the largest van der Waals radius is again the carbon atom. Finally, the distance \(R\) between the geometrical centres of the two molecules is computed as \(R = r_{1} + 2r_{1,vdW} + r_{2} + 2r_{2,vdW}\).
A Schematic representation of the definition of the initial distance between molecules. B Total interaction energy and EDA along five different intermolecular distances and, C polarisation density at the energy minimum for the complex formed by the glycine amino acid and 3-(2-methoxyphenoxy)propane-1,2-diol molecule
Then, for the previously determined intermolecular distance, both molecules are randomly rotated until obtaining 15 different orientations. For each orientation, a single point energy at PM6 level of theory (low level) using the Gaussian09 software [45] has been performed, and the lowest-energy orientation is chosen (step ii). Therefore, by the combination of the steps (i) and (ii), a geometry of the complex that is likely very close to the global minimum of the low-level method is chosen. For this favourable geometry, the potential energy curve is computed at PM6 level in the step (iii) by performing energy calculations at different intermolecular distances, from which the energy of the isolated molecules is subtracted to obtain the interaction energy. Specifically, the intermolecular distance is shortened and elongated in steps of 0.1 Å until the interaction energy is higher than 20 kcal/mol, in the repulsive region, and smaller than 10% of the absolute value of the interaction energy at the energy minimum, in the region of large intermolecular distances. In this way, it is assured that the PM6 potential energy curve considers the intermolecular distances around the energy minimum, which could be thermally populated in a dynamic scenario because they present a favourable, or at least not highly repulsive, interaction energy.
Once the potential energy curve is computed at low level of theory, the interaction energy for five geometries around the low-level energy minimum is refined by higher-level computations. Specifically, the low-level energy minimum geometry, two consecutive geometries at shorter distances, and two geometries taken every two steps at larger distances are selected from the PM6 curve. For those geometries, the input files for computing the interaction energy, step (iv), with the M06-2X functional and 6-31G* basis set using the Gaussian09 software [45] are created with the MoBioTools kit [46, 47]. In the interaction energy calculation, the basis set superposition error is corrected by applying the counterpoise correction [48]. In this first stage of the code development, we aimed at obtaining energetically favourable complex orientations. However, an appropriate training of a force field should also include some repulsive orientations to represent energetically unfavourable regions of the potential energy surface that can be populated along the dynamics. Finally, in the step (v), the electron densities of the complex and the isolated molecules are read by the EDA-NCI program [41, 42, 49] to obtain the different interaction energy contributions, namely, Pauli, electrostatic, dispersion and induction interactions. The sum of the last two terms constitutes the polarisation energy.
The energy contributions are represented in Fig. 2B for the five intermolecular distances of the example complex. As can be seen, for this particular case, the electrostatic, polarisation, dispersion and induction contributions go to more negative values, while the Pauli repulsion term becomes more positive, when the molecules get closer. Overall, the DFT total interaction energy presents a minimum of − 7.36 kcal/mol at 3.49 Å. In this case, the position of the DFT minimum coincides with that of the PM6 one, although this is not the case for all the complexes investigated. The EDA calculation also provides the electron density polarisation induced by the intermolecular interaction. Thus, the functional groups that modify its electron density because of the interaction can be easily identified. Figure 2C displays the polarisation density for the glycine/3-(2-methoxyphenoxy)propane-1,2-diol complex, where it can be seen that there exist an electron density migration from the amino acid to the alcohol group of the drug. All the steps described above for this sample case have been performed for 250 complexes and the results are shown in the Supporting Information.
3 Results and discussion
3.1 Interaction energy and energy decomposition analysis
In this section, the total interaction energy and its repulsive (Pauli) and attractive (electrostatic and polarisation) contributions will be discussed. Figure 3 shows the distributions for the total interaction energy and Pauli repulsion for the 250 complexes. As can be seen in Fig. 3A, the interaction energy distribution is relatively broad, going from − 20 to 3 kcal/mol with an average value of − 4.91 kcal/mol. Since the complexes are formed by different drugs interacting with the same amino acid (glycine), the broadness of the distribution highlights the wide variety of drugs, with functional groups of different nature, including in our set. The chemical structure of the drugs and its possible relation with the EDA will be discussed in the next section. More interestingly, there are only 11 out of 250 molecules that present a positive total interaction energy. This indicates that the procedure driven by the developed python script provides, in most of the cases investigated here, a relative orientation and intermolecular distance that lie on an energetically favourable and, thus, relevant region of the potential energy surface. It is important to notice that the approach of choosing the initial distance between the molecules as the sum of bounding spheres will work properly when the molecules have a nearly spherical symmetry. However, this approach can fail when dealing with linear and large molecules, for which a different methodology must be implemented. For example, doing the random rotations for different initial intermolecular distances (instead of only for one) could be an option, although it would increase the computational time. A different approach could be the random exploration of the molecular surface of one species by the other species as it is done in docking or Monte Carlo simulations. The Pauli repulsion, depicted in Fig. 3B, also presents a large broadness (from 0 to 30 kcal/mol) with an average value of 9.14 kcal/mol.
The distributions of the attractive energy terms, namely, electrostatic, dispersion and induction, are plotted in Fig. 4. Specifically, the distributions are represented in terms of the percentage of each of these three attractive contributions with respect to the total attractive energy. As can be observed, most of the systems have a percentage of electrostatic contribution (Fig. 4A) between 55% and 75%, with a maximum value at 64% and an average value of 59%. Regarding polarisation, the dispersion term (Fig. 4B) represents around 20–40% of the attractive energy with a maximum at 24% and an average value around 29%, while the induction energy contribution (Fig. 4C) is found within 5–20% of the total attraction, with maximum and average values of 8% and 12%, respectively. From this analysis, one can conclude that most of the drug/glycine systems have electrostatic interactions slightly stronger than polarisation ones. However, the polarisation contribution is quite significant, especially dispersion, a fact that is relevant since polarisation is usually hard to describe by conventional fixed-charged force fields, which are very commonly employed in the modelling of drug/protein interactions.
3.2 Energy/structure analysis
In order to analyse whether there exists a relation between the chemical structure of the drugs and the distribution of the interaction energy terms discussed above, different analysis have been carried out. First, the complexes with percentage values of electrostatic, dispersion and induction contributions higher than an arbitrary large threshold have been selected and visualised. This qualitative analysis aimed at finding an evident feature common to all the molecules with a large specific energetic contribution, which can be identified by simple visualization. Specifically, it was found that 27 systems have an electrostatic contribution higher than 70%, 36 systems have the dispersion percentage higher than 40%, and 53 systems have the induction percentage higher than 15%. When each of those sets of systems were visually inspected, no similar structural features were identified among the molecules with a high contribution of the same energetic term. Therefore, the percentage thresholds were increased until only 5 systems per energy term satisfy the newly chosen tighter criterion, resulting in percentages of 87%, 53% and 22% for electrostatic, dispersion and induction, respectively. However, once again no common features among the molecules were evident by visualization.
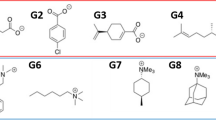
After visualization, a more quantitative and automatic structure-energy analysis was carried out with the goal of finding a potential relation between the presence of some specific functional groups and the dominant attractive energy term in the interaction energy. We have selected eight of the most common functional polar groups presented in bioactive molecules [50] and four aromatic rings common in the set of drugs investigated here. All these functional groups are shown in Fig. 5A, where \(R\) means any aliphatic or aromatic carbon. The functional groups 9–12 in Fig. 5A are not attached to R because they can be bonded to other atoms different from C at different positions of the ring. The first step of this analysis was to represent the different functional groups in the SMILES format using the OpenBabel [51] software. Then, the RDKit python module [52] was applied to check whether a functional group is present in the set of drugs or not, and in how many molecules it is present. The percentage of complexes in our set of systems that possesses each of the functional groups is represented in Fig. 5B. The most frequent functional groups in the drugs investigated here are alcohols (group 7) and carboxyl acids (group 8), which are present in around 80% of the investigated drugs, followed by secondary amines (group 5) with 56%. Next, tertiary amines (group 3) and ethers (group 2) are found with 35% and 33% of occurrence, respectively. All the other functional groups are present in the set of complexes with an occurrence lower than 10%.
Once the most frequent functional groups are identified, the average percentage of each attractive energy contribution (electrostatic, dispersion and induction) with respect to the total attractive energy is computed for the sets of systems that present each of those functional groups. The result is represented in Fig. 5C, where it can be observed that the predominant term is the electrostatic contribution independently on the functional group present in the drug. This can be rationalized by attending to the fact that all the functional groups have polar character, with bonds involving atoms of different electronegativity able to form strong interactions between their permanent charge distributions. Although the percentage of electrostatic contribution is similar along all the sets of molecules, the functional group with the highest electrostatic energy is the thiazole one (group 11). This is a functional group composed by a small ring with large permanent charges due to the presence of a nitrogen and a sulphur atom, which are capable of participating in strong electrostatic interactions. In the case of the polarisation energy (the sum of dispersion and induction), the drugs with the largest contribution are the ones that possess a chlorine atom (group 6). This is not surprising since this atom is easily polarisable due to its large size and, therefore, it can participate in polarisation interactions.
According to the previous analysis, it seems that the presence of small size heteroatoms leads to electrostatic interactions, while large size heteroatoms contribute to the formation of polarisation interactions. To corroborate this, the average of each attractive contribution is computed for the set of drugs that present a specific number of heteroatoms different from chlorine (Fig. 6A) and a specific number of chlorine atoms (Fig. 6B). When the number of heteroatoms (no chlorine) increases, the electrostatic energy term increases (from around 55 to 65%), while the dispersion contribution decreases, and the induction term is kept virtually constant. On the contrary, the electrostatic contribution largely decreases from 60 to 35% when the number of chlorine atoms goes from 0 to 3, while the polarisation term (sum of dispersion and induction) increases. Therefore, the nature and the number of heteroatoms present in the drug determine the importance of the different energetic contributions that control the interaction energy between the drug and the amino acid. However, a much larger set of drugs interacting with additional amino acids is required to generalize these conclusions.
4 Conclusions
Proteins are involved in many key processes in our organism, including the interaction with drugs aimed at treating a wide amount of disorders. Therefore, the study of drug/protein interactions from a computational perspective is crucial to understand the mode of action and side effects of those drugs and develop new ones with improved properties. In this study, we have developed a python code to automatically characterize the interaction within large sets of complexes formed by different drugs and amino acids.
Specifically, the code is able to find an energetically favourable orientation between the two molecules of the complex based on low-level energy calculations. Then, it generates the input files for performing higher-level quantum mechanical calculations of the interaction energy and subsequent EDA to obtain the different energy contributions: Pauli repulsion, electrostatic, dispersion, and induction energies. These calculations are evolved for geometries found around the low-level energy minimum. We have applied the code to characterize the interaction between the glycine amino acid and 250 drugs randomly selected from a database. The broad energy distributions obtained from the analysis indicate that the set of drugs investigated is very diverse, with a wide variety of structures and functional groups. The electrostatic contribution dominates the interaction of most of the drug/glycine systems, although the polarisation terms are also relevant, especially the dispersion one.
The relation between the presence of specific functional groups in the drug structure and the type of energy term that dominates the drug/amino acid interaction was analysed. A clear trend could not be drawn since the electrostatic contribution was found to be dominant independently on the nature of the functional groups. However, the number and size of the heteroatoms correlate well with the nature of the interaction. Thus, the electrostatic and polarisation energies increase when the number of small and large size heteroatoms is increased, respectively. A greater number of calculations, including additional drugs and amino acids, is required to find a more robust and general structure-energy relationship.
References
Wang YC, Zhang CH, Deng NY, Wang Y (2011) Kernel-based data fusion improves the drug-protein interaction prediction. Comput Biol Chem 35:353–362. https://doi.org/10.1016/j.compbiolchem.2011.10.003
Lombardi D, Dittrich PS (2011) Droplet microfluidics with magnetic beads: A new tool to investigate drug-protein interactions. Anal Bioanal Chem 399:347–352. https://doi.org/10.1007/s00216-010-4302-7
Hage DS, Jackson A, Sobansky MR, Schiel JE, Yoo MJ, Joseph KS (2009) Characterization of drug-protein interactions in blood using high-performance affinity chromatography. J Sep Sci 32:835–853. https://doi.org/10.1002/jssc.200800640
Zheng S, Li Y, Chen S, Xu J, Yang Y (2020) Predicting drug-protein interaction using quasi-visual question answering system. Nat Mach Intell 2:134–140. https://doi.org/10.1038/s42256-020-0152-y
Sharma H, Navalkar A, Maji SK, Agrawal A (2019) Analysis of drug-protein interaction in bio-inspired microwells. SN Appl Sci 1:819. https://doi.org/10.1007/s42452-019-0778-8
Ding F, Dokholyan NV, Buldyrev SV, Stanley HE, Shakhnovich EI (2002) Direct molecular dynamics observation of protein folding transition state ensemble. Biophys J 83:3525–3532. https://doi.org/10.1016/S0006-3495(02)75352-6
Daggett V (2002) Molecular dynamics simulations of the protein unfolding/folding reaction. Acc Chem Res 35:422–429. https://doi.org/10.1021/ar0100834
Gsponer J, Caflisch A (2002) Molecular dynamics simulations of protein folding from the transition state. Proc Natl Acad Sci USA 99:6719–6724. https://doi.org/10.1073/pnas.092686399
Scheraga HA, Khalili M, Liwo A (2007) Protein-folding dynamics: overview of molecular simulation techniques. Annu Rev Phys Chem 58:57–83. https://doi.org/10.1146/annurev.physchem.58.032806.104614
Durrant JD, McCammon JA (2011) Molecular dynamics simulations and drug discovery. BMC Biol 9:71. https://doi.org/10.1186/1741-7007-9-71
De Vivo M, Masetti M, Bottegoni G, Cavalli A (2016) Role of molecular dynamics and related methods in drug discovery. J Med Chem 59:4035–4061. https://doi.org/10.1021/acs.jmedchem.5b01684
Legina MS, Nogueira JJ, Kandioller W, Jakupec MA, González L, Keppler BK (2020) Biological evaluation of novel thiomaltol-based organometallic complexes as topoisomerase IIα Inhibitors. J Biol Inorg Chem 25:451–465. https://doi.org/10.1007/s00775-020-01775-2
Szabó PB, Sabanés Zariquiey F, Nogueira JJ (2021) Cosolvent and dynamic effects in binding pocket search by docking simulations. J Chem Inf Model 61:5508–5523. https://doi.org/10.1021/acs.jcim.1c00924
Noh SY, Notman R (2020) Comparison of umbrella sampling and steered molecular dynamics methods for computing free energy profiles of aromatic substrates through phospholipid bilayers. J Chem Phys 153:034115. https://doi.org/10.1063/5.0016114
Lee CT, Comer J, Herndon C, Leung N, Pavlova A, Swift RV, Tung C, Rowley CN, Amaro RE, Chipot C, Wang Y, Gumbart JC (2016) Simulation-based approaches for determining membrane permeability of small compounds. J Chem Inf Model 56:721–733. https://doi.org/10.1021/acs.jcim.6b00022
Neale C, Pomès R (1858) Sampling errors in free energy simulations of small molecules in lipid bilayers. Biochim Biophys Acta Biomembr 2016:2539–2548. https://doi.org/10.1016/j.bbamem.2016.03.006
Nogueira JJ, Meixner M, Bittermann M, González L (2017) Impact of lipid environment on photodamage activation of methylene blue. ChemPhotoChem 1:178–182. https://doi.org/10.1002/cptc.201600062
Sánchez-Murcia PA, Nogueira JJ, González L (2018) Exciton localization on Ru-based photosensitizers induced by binding to lipid membranes. J Phys Chem Lett 9:683–688. https://doi.org/10.1021/acs.jpclett.7b03357
Ruano L, Cárdenas G, Nogueira JJ (2021) The permeation mechanism of cisplatin through a dioleoylphosphocholine bilayer**. ChemPhysChem 22:1251–1261. https://doi.org/10.1002/cphc.202100059
Law YK, Azadi J, Crespo-Hernández CE, Olmon E, Kohler B (2008) Predicting thymine dimerization yields from molecular dynamics simulations. Biophys J 94:3590–3600. https://doi.org/10.1529/biophysj.107.118612
Bignon E, Claerbout VEP, Jiang T, Morell C, Gillet N, Dumont E (2020) Nucleosomal embedding reshapes the dynamics of abasic sites. Sci Rep 10:17314. https://doi.org/10.1038/s41598-020-73997-y
Abolfath RM, Carlson DJ, Chen ZJ, Nath R (2013) A molecular dynamics simulation of DNA damage induction by ionizing radiation. Phys Med Biol 58:7143–7157. https://doi.org/10.1088/0031-9155/58/20/7143
Nogueira JJ, González L (2014) Molecular dynamics simulations of binding modes between methylene blue and DNA with alternating GC and AT sequences. Biochemistry 53:2391–2412. https://doi.org/10.1021/bi500068z
Czapla-Masztafiak J, Nogueira JJ, Lipiec E, Kwiatek WM, Wood BR, Deacon GB, Kayser Y, Fernandes DLA, Pavliuk MV, Szlachetko J, González L, Sá J (2017) Direct determination of metal complexes’ interaction with DNA by atomic telemetry and multiscale molecular dynamics. J Phys Chem Lett 8:805–811. https://doi.org/10.1021/acs.jpclett.7b00070
Sotomayor M, Vásquez V, Perozo E, Schulten K (2007) Ion Conduction through MscS as determined by electrophysiology and simulation. Biophys J 92:886–902. https://doi.org/10.1529/biophysj.106.095232
Kopec W, Köpfer DA, Vickery ON, Bondarenko AS, Jansen TLC, de Groot BL, Zachariae U (2018) Direct Knock-on of desolvated ions governs strict ion selectivity in K+ channels. Nat Chem 10:813–820. https://doi.org/10.1038/s41557-018-0105-9
Ulmschneider MB, Bagnéris C, McCusker EC, DeCaen PG, Delling M, Clapham DE, Ulmschneider JP, Wallace BA (2013) Molecular dynamics of ion transport through the open conformation of a bacterial voltage-gated sodium channel. Proc Natl Acad Sci USA 110:6364–6369. https://doi.org/10.1073/pnas.1214667110
Li Y, Sun R, Liu H, Gong H (1859) Molecular dynamics study of ion transport through an open model of voltage-gated sodium channel. Biochim Biophys Acta Biomembr 2017:879–887. https://doi.org/10.1016/j.bbamem.2017.02.003
Oakes V, Furini S, Domene C (2016) Voltage-gated sodium channels: mechanistic insights from atomistic molecular dynamics simulations. Curr Top Membr 78:183–214. https://doi.org/10.1016/bs.ctm.2015.12.002
Corry B, Thomas M (2012) Mechanism of Ion permeation and selectivity in a voltage gated sodium channel. J Am Chem Soc 134:1840–1846. https://doi.org/10.1021/ja210020h
Ponder JW, Case DA (2003) Force fields for protein simulations. Adv Protein Chem 66:27–85. https://doi.org/10.1016/S0065-3233(03)66002-X
Huang J, Rauscher S, Nawrocki G, Ran T, Feig M, De Groot BL, Grubmüller H, MacKerell AD (2016) CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nat Methods 14:71–73. https://doi.org/10.1038/nmeth.4067
Jorgensen WL, Maxwell DS, Tirado-Rives J (1996) Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J Am Chem Soc 118:11225–11236. https://doi.org/10.1021/ja9621760
González MA (2011) Force fields and molecular dynamics simulations. J de La Neutronique 12:169–200. https://doi.org/10.1051/sfn/201112009
Chmiela S, Sauceda HE, Müller KR, Tkatchenko A (2018) Towards exact molecular dynamics simulations with machine-learned force fields. Nat Commun 9:3887. https://doi.org/10.1038/s41467-018-06169-2
Botu V, Batra R, Chapman J, Ramprasad R (2017) Machine learning force fields: construction, validation, and outlook. J Phys Chem C 121:511–522. https://doi.org/10.1021/acs.jpcc.6b10908
Unke OT, Chmiela S, Sauceda HE, Gastegger M, Poltavsky I, Schütt KT, Tkatchenko A, Müller KR (2021) machine learning force fields. Chem Rev 121:10142–10186. https://doi.org/10.1021/acs.chemrev.0c01111
Handley CM, Popelier PLA (2010) Potential energy surfaces fitted by artificial neural networks. J Phys Chem A 114:3371–3383. https://doi.org/10.1021/jp9105585
Behler J (2017) First principles neural network potentials for reactive simulations of large molecular and condensed systems. Angew Chem Int Ed 56:12828–12840. https://doi.org/10.1002/anie.201703114
Metcalf DP, Koutsoukas A, Spronk SA, Claus BL, Loughney DA, Johnson SR, Cheney DL, Sherrill CD (2020) Approaches for machine learning intermolecular interaction energies and application to energy components from symmetry adapted perturbation theory. J Chem Phys 152:074103. https://doi.org/10.1063/1.5142636
Mandado M, Hermida-Ramón JM (2011) Electron density based partitioning scheme of interaction energies. J Chem Theory Comput 7:633–641. https://doi.org/10.1021/ct100730a
Ramos-Berdullas N, Pérez-Juste I, van Alsenoy C, Mandado M (2015) Theoretical study of the adsorption of aromatic units on carbon allotropes including explicit (empirical) DFT dispersion corrections and implicitly dispersion-corrected functionals: the pyridine case. Phys Chem Chem Phys 17:575–587. https://doi.org/10.1039/c4cp02341b
Sterling T, Irwin JJ (2015) ZINC 15: ligand discovery for everyone. J Chem Inf Model 55:2324–2337. https://doi.org/10.1021/acs.jcim.5b00559
Ropo M, Schneider M, Baldauf C, Blum V (2016) First-principles data set of 45,892 isolated and cation-coordinated conformers of 20 proteinogenic amino acids. Sci Data 3:160009. https://doi.org/10.1038/sdata.2016.9
Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Scalmani G, Barone V, Petersson GA, Nakatsuji H, Li X, Caricato M, Marenich AV, Bloino J, Janesko BG, Gomperts R, Mennucci B, HP Hratchian, Ortiz JV, Izmaylov AF, Sonnenberg JL, Williams, Ding F, Lipparini F, Egidi F, Goings J, Peng B, Petrone A, Henderson T, Ranasinghe D, Zakrzewski VG, Gao J, Rega N, Zheng G, Liang W, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Vreven T, Throssell K, Montgomery Jr JA, Peralta JE, Ogliaro F, Bearpark MJ, Heyd JJ, Brothers EN, Kudin KN, Staroverov VN, Keith TA, Kobayashi R, Normand J, Raghavachari K, Rendell AP, Burant JC, Iyengar SS, Tomasi J, Cossi M, Millam JM, Klene M, Adamo C, Cammi R, Ochterski JW, Martin RL, Morokuma K, Farkas O, Foresman JB, Fox DJ (2016) Gaussian 09
Cárdenas G, Lucia-Tamudo J, Mateo-delaFuente H, Palmisano VF, Anguita-Ortiz N, Ruano L, Pérez-Barcia Á, Díaz-Tendero S, Mandado M, Nogueira JJ (2023) MoBioTools: a toolkit to setup quantum mechanics/molecular mechanics calculations. J Comput Chem 44:516–533. https://doi.org/10.1002/jcc.27018
Cárdenas G, Nogueira JJ (2022) MoBioTools. https://github.com/mobiochem/MoBioTools
Boys SF, Bernardi F (1970) The calculation of small molecular interactions by the differences of separate total energies: some procedures with reduced errors. Mol Phys 100:65–73. https://doi.org/10.1080/00268970110088901
Mandado M, van Alsenoy C (n.d.) EDA-NCI. https://github.com/marcos-mandado/EDA-NCI
Ertl P, Altmann E, McKenna JM (2020) The most common functional groups in bioactive molecules and how their popularity has evolved over time. J Med Chem 63:8408–8418. https://doi.org/10.1021/acs.jmedchem.0c00754
O’Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR (2011) Open babel: an open chemical toolbox. J Cheminform 3:33. https://doi.org/10.1186/1758-2946-3-33
RDKit: open-source cheminformatics., (n.d.). https://www.rdkit.org
Acknowledgements
LR and JJN acknowledge the Comunidad de Madrid for funding through the Attraction of Talent Program (Grant ref 2018-T1/BMD-10261) and the Spanish Ministry of Science and Innovation (Project PID2020-117806GA-I00). MM thanks Xunta de Galicia for financial support through the project GRC2019/24. We are also grateful for the generous allocation of computer time and continued technical support by the Centro de Computación Científica of the UAM (CCC-UAM)
Funding
Open Access funding provided thanks to the CRUE-CSIC agreement with Springer Nature.
Author information
Authors and Affiliations
Contributions
LR: Data curation, formal analysis, investigation, software, validation, visualization, writing original draft, writing—review and editing. MM and JJN: Conceptualization, funding acquisition, project administration, resources, supervision, writing—review and editing.
Corresponding authors
Ethics declarations
Conflict of interest
The authors have no competing interests to declare.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ruano, L., Mandado, M. & Nogueira, J.J. Automatic characterization of drug/amino acid interactions by energy decomposition analysis. Theor Chem Acc 142, 55 (2023). https://doi.org/10.1007/s00214-023-02997-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00214-023-02997-8