Abstract
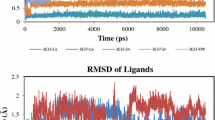
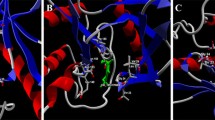
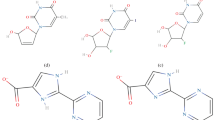
FK506-binding protein (FKBP) is a cytosolic enzyme, which catalyzes the cis–trans isomerization of prolyl peptides. The twisted amide geometries assist the isomerization process and inhibit the rotamase activity of FKBP. This work reveals the role of electronic effects in ligands to facilitate the twisted amide geometries for efficient inhibition of the rotamase activity of FKBP12. The azirine ligand (5A) lowered the rotation barrier of –C–N amide bond by ~ 14.0 kcal/mol compared to the reference ligand molecule (1A) using M062X/cc-pVDZ//M062X/6-31G(d) level of theory. A significant lowering in rotation barrier has been achieved in (5A) due to the ring's anti-aromatic nature. The ring strain in the 1H-azirine ring pyramidalizes the amide bond's nitrogen center by 78° in the ground state compared to 1A. The natural bond orbital (NBO) analysis confirmed the lowering in the delocalization of electron density from nitrogen lone pair to π*C = O orbital in amide bonds. The designed models (1A-5A) were developed into ligands to mimic the binding domain of FK506 (1B-5B). The binding of these ligands (1B-5B) was examined with the quantum mechanics and molecular mechanics (QM/MM) ONIOM approach in the presence of FKBP12 binding site residues. The calculated results reveal the preferential binding of twisted amide forms with FKBP12 over their planar analogous. The hydrophobic interactions augment the binding affinity of the twisted forms with the FKBP12 binding site in agreement with the experimental observations. These studied ligands could act as potential inhibitors of FKBP12 for their therapeutic application.





Similar content being viewed by others
Abbreviations
- FKBP:
-
FK506-binding proteins
- DFT:
-
Density functional theory
- QM/MM:
-
Quantum mechanics and molecular mechanics
- TS:
-
Transition state
- GS:
-
Ground state
- NBO:
-
Natural bond orbital
- PDB:
-
Protein Data Bank
References
Kolos JM, Voll AM, Bauder M, Hausch F (2018) FKBP ligands—where we are and where to go? Front Pharmacol 9:1425. https://doi.org/10.3389/fphar.2018.01425
Wiederrecht G, Hung S, Chan HK et al (1992) Characterization of high molecular weight FK-506 binding activities reveals a novel FK-506-binding protein as well as a protein complex. J Biol Chem 267:21753–60
Orozco M, Tirado-Rives J, Jorgensen WL (1993) Mechanism for the rotamase activity of FK506 binding protein from molecular dynamics simulations. Biochemistry 32:12864–12874. https://doi.org/10.1021/bi00210a040
Rosen M, Standaert R, Galat A et al (1990) Inhibition of FKBP rotamase activity by immunosuppressant FK506: twisted amide surrogate. Science 248:863–866. https://doi.org/10.1126/science.1693013
Park ST, Aldape RA, Futer O et al (1992) PPIase catalysis by human FK506-binding protein proceeds through a conformational twist mechanism. J Biol Chem 267:3316–3324
Fischer S, Michnick S, Karplus M (1993) A mechanism for rotamase catalysis by the FK506 binding protein (FKBP). Biochemistry 32:13830–13837. https://doi.org/10.1021/bi00213a011
Albers MW, Walsh CT, Schreiber SL (1990) Substrate specificity for the human rotamase FKBP: a view of FK506 and rapamycin as leucine-(twisted amide)-proline mimics. J Org Chem 55:4984–4986. https://doi.org/10.1021/jo00304a003
Itoh S, DeCenzo MT, Livingston DJ et al (1995) Conformation of FK506 in X-ray structures of its complexes with human recombinant FKBP12 mutants. Bioorganic Med Chem Lett 5:1983–1988. https://doi.org/10.1016/0960-894X(95)00337-S
Van Duyne G, Standaert R, Karplus P et al (1991) Atomic structure of FKBP-FK506, an immunophilin-immunosuppressant complex. Science 252:839–842. https://doi.org/10.1126/science.1709302
Kesharwani MK, Ganguly B (2011) Probing the structural and electronic effects to stabilize nonplanar forms of thioamide derivatives: a computational study. J Comput Chem 32:2170–2176. https://doi.org/10.1002/jcc.21800
Limburg DC, Thomas BE, Li J et al (2003) Synthesis and evaluation of chiral bicyclic proline FKBP12 ligands. Bioorg Med Chem Lett 13:3867–3870. https://doi.org/10.1016/S0960-894X(03)00758-3
Bischoff M, Sippel C, Bracher A, Hausch F (2014) Stereoselective construction of the 5-hydroxy diazabicyclo[4.3.1]decane-2-one scaffold, a privileged motif for FK506-binding proteins. Org Lett 16:5254–5257. https://doi.org/10.1021/ol5023195
Dunyak BM, Gestwicki JE (2016) Peptidyl-proline isomerases (PPIases): targets for natural products and natural product-inspired compounds. J Med Chem 59:9622–9644. https://doi.org/10.1021/acs.jmedchem.6b00411
Pomplun S, Wang Y, Kirschner A et al (2015) Rational design and asymmetric synthesis of potent and neurotrophic ligands for FK506-binding proteins (FKBPs). Angew Chemie Int Ed 54:345–348
Beom JY, Jung JA, Lee K et al (2019) Biosynthesis of nonimmunosuppressive FK506 analogues with antifungal activity. J Nat Prod 82:2078–2086. https://doi.org/10.1021/acs.jnatprod.9b00144
Pomplun S, Sippel C, Hähle A et al (2018) Chemogenomic profiling of human and microbial FK506-binding proteins. J Med Chem 61:3660–3673. https://doi.org/10.1021/acs.jmedchem.8b00137
Hamilton GS, Wu Y, Limburg DC et al (2002) Synthesis of N-glyoxyl prolyl and pipecolyl amides and thioesters and evaluation of their in vitro and in vivo nerve regenerative effects. J Med Chem 45:3549–3557. https://doi.org/10.1021/jm010556c
Hamilton GS, Huang W, Connolly MA et al (1997) FKBP12-binding domain analogues of FK506 are potent, nonimmunosuppressive neurotrophic agents in vitro and promote recovery in a mouse model of parkinson’s disease. Bioorg Med Chem Lett 7:1785–1790. https://doi.org/10.1016/S0960-894X(97)00304-1
Hur S, Bruice TC (2002) The mechanism of cis-trans isomerization of prolyl peptides by cyclophilin. J Am Chem Soc 124:7303–7313. https://doi.org/10.1021/ja020222s
Zhang H, Zheng Q (2019) How chorismatases regulate distinct reaction channels in a single conserved active pocket? Revealed by mechanistic analysis with QM/MM (ONIOM) investigations Yulai. Chem Eur J 25:1326–1336. https://doi.org/10.1002/chem.201804622
Jayasinghe-arachchige VM, Hu Q, Sharma G et al (2018) Hydrolysis of chemically distinct sites of human serum albumin by polyoxometalate : a hybrid QM/MM (ONIOM) study. J Comput Chem 40:51–61. https://doi.org/10.1002/jcc.25528
Fernandes HS, Ramos MJ, Cerqueira NM (2018) The catalytic mechanism of the serine hydroxymethyltransferase—a computational ONIOM QM/MM study. ACS Catal 89(11):10096–10110. https://doi.org/10.1021/acscatal.8b02321
Gaussian 09, Revision D.01, Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Scalmani G, Barone V, Mennucci B, Petersson GA et al. (2013) Gaussian, Inc., Wallingford CT
Zhao Y, Truhlar DG (2008) The M06 suite of density functionals for main group thermochemistry, thermochemical kinetics, noncovalent interactions, excited states, and transition elements: two new functionals and systematic testing of four M06-class functionals and 12 other function. Theor Chem Acc 120:215–241. https://doi.org/10.1007/s00214-007-0310-x
Rassolov VA, Ratner MA, Pople JA et al (2001) 6–31G* basis set for third-row atoms. J Comput Chem 22:976–984. https://doi.org/10.1002/jcc.1058
Cossi M, Rega N, Scalmani G, Barone V (2003) Energies, structures, and electronic properties of molecules in solution with the C-PCM solvation model. J Comput Chem 24:669–681. https://doi.org/10.1002/jcc.10189
Wilson KP, Yamashita MM, Sintchak MD et al (1995) Comparative X-ray structures of the major binding protein for the immunosuppressant FK506 (tacrolimus) in unliganded form and in complex with FK506 and rapamycin. Acta Crystallogr Sect D Biol Crystallogr 51:511–521. https://doi.org/10.1107/S0907444994014514
Delano WL (2002) The PyMOL molecular graphics system
Dallakyan S, Olson AJ (2016) Small-molecule library screening by docking with PyRx. Methods Mol Biol 1263:243–50. https://doi.org/10.1007/978-1-4939-2269-7
Li S, Hong M (2011) Protonation, tautomerization, and rotameric structure of histidine: a comprehensive study by magic-angle-spinning solid-state NMR. J Am Chem Soc 133:1534–1544. https://doi.org/10.1021/ja108943n
Reynolds WF, Peat IR, Freedman MH, Lyerla JR Jr (1973) Additions and corrections—determination of the tautomeric form of the imidazole ring of L-histidine in basic solution by carbon-13 magnetic resonance spectroscopy. J Am Chem Soc 95:6510–6511. https://doi.org/10.1021/ja00800a603
Trott O, Olson AJ (2011) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J Comput Chem 31:455–461. https://doi.org/10.1002/jcc.21334.AutoDock
Dewar MJS, Zoebisch EG, Healy EF, Stewart JJP (1985) AM1: A new general purpose quantum mechanical molecular model1. J Am Chem Soc 107:3902–3909. https://doi.org/10.1021/ja00299a024
Martina MR, Tenori E, Bizzarri M et al (2013) The precise chemical-physical nature of the pharmacore in FK506 binding protein inhibition: ElteX, a new class of nanomolar FKBP12 ligands. J Med Chem 56:1041–1051. https://doi.org/10.1021/jm3015052
Szostak R, Szostak M (2019) Chemistry of bridged lactams: recent developments. Molecules 24:274. https://doi.org/10.3390/molecules24020274
Császár AG, Demaison J, Rudolph HD (2015) Equilibrium structures of three-, four-, five-, six-, and seven-membered unsaturated n-containing heterocycles. J Phys Chem A 119:1731–1746. https://doi.org/10.1021/jp5084168
Cheisson T, Manor BC, Carroll PJ et al (2019) C NMR shifts as an indicator of U–C bond covalency in uranium(VI) acetylide complexes: an experimental and computational study ́. Inorg Chem 337:4152–4163. https://doi.org/10.1021/acs.inorgchem.8b03175
Suslonov VV, Luzyanin KV, Kukushkin VY (2017) Tetrazol-5-ylidene gold(III) complexes from sequential [2 + 3] cycloaddition of azide to metal-bound Isocyanides and N4 alkylation. Organometallics 36:3974–3980. https://doi.org/10.1021/acs.organomet.7b00591
Bikbaeva ZM, Novikov AS, Suslonov VV et al (2017) Metal-mediated reactions between dialkylcyanamides and acetamidoxime generate unusual (nitrosoguanidinate)nickel(II) complexes. Dalt Trans 46:10090–10101. https://doi.org/10.1039/c7dt01960b
Bucciarelli M, Forni A, Moretti I et al (1993) Candida cylindracea lipase-catalysed hydrolysis of methyl aziridine-2-carboxylates and -2,3-dicarboxylates. J Chem Soc Perkin Trans 1:3041–3045. https://doi.org/10.1039/p19930003041
Lanter JC, Zhang S, Zhao B (2003) Ortho McNeil Pharmaceutical Inc, Neurotrophic 2-azetidinecarboxylic acid derivatives, and related compositions and methods. U.S. Patent 6,544,976
Meinwald J, Aue DH (1966) The photochemical addition of methyl azidoformate to 2-butyne. J Am Chem Soc 88:2849–2850. https://doi.org/10.1021/ja00964a041
Huisgen R, Blashke H (1965) 2-Äthoxy-oxazole aus Äthoxycarbonyl-azen und Alkinen. Chem Ber 599:2985–2997
Tetko IV, Tanchuk VY, Kasheva TN, Villa AEP (2001) Internet software for the calculation of the lipophilicity and aqueous solubility of chemical compounds. J Chem Inf Comput Sci 41:246–252. https://doi.org/10.1021/ci000393l
Tetko IV, Gasteiger J, Todeschini R et al (2005) Virtual computational chemistry laboratory—design and description. J Comput Aided Mol Des 19:453–463. https://doi.org/10.1007/s10822-005-8694-y
Tetko IV, Villa AEP (1997) Efficient partition of learning data sets for neural network training. Neural Netw 10:1361–1374. https://doi.org/10.1016/S0893-6080(97)00005-1
Kyte J, Doolittle RF (1982) A simple method for displaying the hydropathic character of a protein. J Mol Biol 157:105–132. https://doi.org/10.1016/0022-2836(82)90515-0
Chang KY, Yang J-R (2013) Analysis and prediction of highly effective antiviral peptides based on random forests. PLoS ONE 8:e70166. https://doi.org/10.1371/journal.pone.0070166
Lipinski CA, Lombardo F, Dominy BW, Feeney PJ (2012) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 64:4–17. https://doi.org/10.1016/j.addr.2012.09.019
Lipinski CA (2004) Lead- and drug-like compounds: the rule-of-five revolution. Drug Discov Today Technol 1:337–341. https://doi.org/10.1016/j.ddtec.2004.11.007
Acknowledgements
P. D. W. acknowledges CSIR, New Delhi, India, for the GATE-junior research fellowship, and P. D. W. is thankful to AcSIR Ghaziabad, Uttar Pradesh-201 002, India, for her Ph.D. registration. B.G. thanks Department of Science and Technology, Government of India, under (grant no. EMR/2017/004652) and Department of Biotechnology (Grant no. BT/PR12730/BID/7/523/ 2015), New Delhi, for financial support. We thank the reviewers for their valuable comments and suggestions that have helped us to improve the article.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
There are no conflicts of interest to declare.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
About this article
Cite this article
Wakchaure, P.D., Ganguly, B. Tuning the electronic effects in designing ligands for the inhibition of rotamase activity of FK506 binding protein. Theor Chem Acc 140, 5 (2021). https://doi.org/10.1007/s00214-020-02717-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00214-020-02717-6