Abstract
Many of the expanding roles of nucleoside diphosphate kinase have been attributed to its ability to interact with other proteins. One proposal is an interaction with the cellular energy sensor AMP-activated protein kinase, and here, we apply the simple eukaryotic organism, Dictyostelium discoideum as a test model. Stable cotransformants were created in which NDPK expression was knocked down by antisense inhibition, and AMPK activity was chronically elevated either by constitutive overexpression of its active, catalytic domain (AMPKαT) or as a result of mitochondrial dysfunction (created by antisense inhibition of expression of a mitochondrial chaperone protein, chaperonin 60). To investigate a biochemical interaction, transformants were created which contained constructs expressing FLAG-NDPK and hexahistidine-tagged full-length AMPK or AMPKαT. The protein extract from these transformants was used in coimmunoprecipitations. Knock down of NDPK expression suppressed the phenotypic defects that are caused by AMPK hyperactivity resulting either from overexpression of AMPKαT or from mitochondrial dysfunction. These included rescue of defects in slug phototaxis, fruiting body morphology and growth in a liquid medium. Coimmunoprecipitation experiments failed to demonstrate a biochemical interaction between the two proteins. The results demonstrate a genetic interaction between NDPK and AMPK in Dictyostelium in that NDPK is required for the phenotypic effects of activated AMPK. Coimmunoprecipitations suggest that this interaction is not mediated by a direct interaction between the two proteins.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Ten genes belonging to the NDPK family have been identified in humans (Boissan et al. 2009). Four (NDPK-A–D) belong to group I, all of which display enzymatic activity and share high sequence identity ranging from 58–88% (Lacombe et al. 2000). NDPK-A and NDPK-B are the most abundant and by far the most studied. These two enzymes arose by an ancient gene duplication event, share 88% identity and are mainly cytoplasmic. That they can substitute for one another to serve some essential functions is best illustrated by the neonatal death of animals bearing deletions of both isoforms, whereas the individual deletions are compatible with survival (Postel et al. 2009; Arnaud-Dabernat et al. 2003). Yet, despite their similarities, these two proteins also have distinct cellular functions (Bosnar et al. 2004), and accumulating evidence suggests that this results from different protein–protein interactions. Additionally, in metastasis, the expression level of NDPK-A (and not NDPK-B) rather than the catalytic activity is the important modulating factor as the latter does not correlate with metastasis suppression. Furthermore, apart from the k-pn mutation in Drosophila (Biggs et al. 1988) and the P120S mutation in childhood neuroblastoma (Chang et al. 1994), no mutations to NDPK have been observed in metastatic cells.
Much research has focused on the antimetastatic role of NDPK, but it seems likely that this function is an adaptation of its more basic, evolutionarily conserved role in normal cells. There are many potential benefits of exploiting the tractability of lower eukaryotes in studying these basic cellular functions. Firstly, the cellular machinery of lower organisms is similar to that of mammalian cells, but the structure of the genome, tissues and organs is simpler and more amenable to experimental study. Secondly, there are fewer NDPK isoforms, which facilitates biochemical and genetic studies. The Drosophila example above is a case in point–mutation of the isoform of NDPK that constitutes around 98% of NDPK activity causes abnormal wing discs.
Dictyostelium discoideum is an ideal model eukaryote. It has a haploid genome which has been completely sequenced (Eichinger et al. 2005) and is amenable to a range of genetic manipulation techniques. It also has a unique life cycle with unicellular (amoeboid) and different multicellular stages that provide an unparalleled variety of phenotypes for cell biological study. As such, Dictyostelium has become a widely accepted model for the study of many processes including mitochondrial disease (Kotsifas et al. 2002; Wilczynska et al. 1997; Bokko et al. 2007). D. discoideum contains three genes encoding NDPK or NDPK-related proteins. A cytosolic form of NDPK, which represents most of the NDPK activity within these cells, is encoded by ndkC (or gip17) (Lacombe et al. 1990) and is the Dictyostelium homologue of human cytosolic group I NDPKs. It was this Dictyostelium NDPK which was used to determine the first NDPK crystal structure and to conduct the first structure–function studies using mutant forms of the enzyme (Lacombe et al. 1990; Dumas et al. 1992; Karlsson et al. 1996; Morera et al. 1994a, b, 1995; Cherflis et al. 1994; Annesley and Fisher 2009).
The involvement of NDPK in many functions has shown to be independent of its catalytic activity, and hence, many speculate that these roles are facilitated by its ability to interact with other proteins. One such interaction that has been reported to occur in epithelial cells of the upper respiratory tract is with the cellular energy sensor protein AMP-activated protein kinase (AMPK) (Muimo et al. 2006; Treharne et al. 2009). Although it has not yet been confirmed for other cell types, such an interaction seems plausible and could be widespread because both NDPK and AMPK play regulatory roles in a number of common cellular functions. These include cell growth and proliferation, fat metabolism (ATP-citrate lyase, the primary source of cytosolic acetyl CoA, regulated by NDPK (Wagner and Vu 1995) and acetyl CoA carboxylase, regulated by AMPK (Hardie 1992)) and protection against oxidative stress (Arnaud-Dabernat et al. 2004; Onken and Driscoll 2010). AMPK acts as a cellular alarm by sensing low energy levels and responding to this by inhibiting energy-consuming pathways and activating energy-producing pathways. This cellular alarm is extremely sensitive and exerts its effects prior to a serious depletion in energy, thereby allowing it to play a central role in cellular energy homeostasis. We reported that AMPK is chronically activated in mitochondrial disease, and this mediates diverse cytopathologies caused by mitochondrial dysfunction (Bokko et al. 2007; Kotsifas et al. 2002). To investigate whether NdkC (henceforth, simply called NDPK) interacts with AMPK in these pathways, genetic and biochemical experiments were performed. A genetic interaction was revealed by the fact that antisense-inhibiting NDPK expression suppressed the phenotypic defects that are otherwise caused by chronic AMPK hyperactivity.
Methods
Dictyostelium strains and culture conditions
Dictyostelium wild-type strain, AX2, and the derived transformants were either grown axenically in HL-5 medium (Watts and Ashworth 1970) which, for transformed cell lines, contained 20 μg/ml G418 (Promega, Annandale, Australia) in shaken suspension (150 rpm) at 21°C or on SM agar plates with Klebsiella aerogenes as a food source (Sussman 1966). The transformants used for genetic interaction experiments included NDPK antisense-inhibited strains (containing construct pPROF500), NDPK overexpressing strains (containing construct pPROF520), AMPKαT overexpressing strains (containing construct pPROF392 (Bokko et al. 2007)), cotransformants of NDPK antisense-inhibited/chaperonin 60 antisense-inhibited strains (containing constructs pPROF500 and pPROF128 (Kotsifas et al. 2002)) and AMPK overexpressing/NDPK antisense-inhibited strains (containing constructs pPROF392 and pPROF500). For coimmunoprecipitation experiments, we used stable cotransformants expressing epitope-tagged forms of NDPK and AMPK. These were FLAG-tagged NDPK (expressed from construct pPROF587) in combination with either hexahistidine-tagged full-length AMPKα (expressed from construct pPROF589) or hexahistidine-tagged AMPKαT (expressed from construct pPROF588).
Plasmid construction
The full-length NDPK gene was amplified using genomic AX2 DNA as template and primers NDPKF (ATCTGAGAATTCATCGATATGTCCACAAATAAAGTAAA) and NDPKR (ATCTGAGAATTCCTCGAGTTATTCGTATAAATTTGGGT).
The PCR product was cloned into the EcoRI site of an Escherichia coli vector (pUC18) and subcloned into the EcoRI site of the Dictyostelium shuttle vector pDNeo2 in the antisense orientation to produce construct pPROF500 for use in antisense inhibition experiments.
The AMPKα full-length gene was amplified from genomic AX2 DNA using the primers “AMPK,FL/F” (GATAGTCTAGATTCGAAATGCATCATCATCATCATCATAGTTCATATCAAC) and “AMPK,FL/R” (CGTATGTCTAGACTCGAGTTATTATAAATTCAATTCCACC). AMPKαT was amplified using pPROF392 as template (containing the cDNA sequence, (Bokko et al. 2007)) and the primers “ampk,his,Ctrunc/F” (GATAGTCTAGATTCGAAATGCATCATCATCATCATCATAGTTCATATCAAC) and “ampk,his,Ctrunc/R” (GTATGTCTAGACTCGAGTTATTATGGACTCTTTTGAGCACTTGACATAATTGG). Both hexahistidine-tagged products were digested with SfuI and XhoI and cloned into the Dictyostelium expression vector pA15GFP (digested with ClaI and XhoI), replacing the GFP gene to produce the constructs pPROF589 (his-AMPKα full length) and pPROF588 (his-AMPKαT). The FLAG-tagged NdkC construct was made by amplifying the NdkC gene using pPROF519 as template and the primers “Flag, ndkc/F” (GATAGGAATTCATCGATATGGACTACAAGGACGATGACAAAATGTCCAC) and “Flag, ndkc/R” (GTATGGAATTCCTCGAGTTATTATTCGTATAAATTTGGGTTTGG). The PCR products were digested with ClaI and XhoI and cloned into the Dictyostelium expression vector pA15GFP, replacing the GFP gene to produce the construct pPROF587.
Transformation
AX2 cells were transformed using 20 μg of each of the relevant constructs with the calcium phosphate DNA precipitation method (Nellen et al. 1984). Following selection on Micrococcus luteus lawns grown on SM agar plates containing 20 μg/ml G418 (Wilczynska and Fisher 1994), transformants were subcultured and maintained on K. aerogenes lawns and axenically in HL-5.
Calculation of construct copy numbers
Genomic DNA was extracted from cells using DNAzol (Molecular Research Center, CI, Ohio) and the instructions provided by the manufacturer. Half of the genomic DNA sample was digested with enzymes to release a band containing the endogenous gene and also to release the gene from the construct. The signals were detected using a fluorescently labelled probe for the fragment of interest in combination with the enhanced chemiluminescence system (ECL, Amersham). The other half of the sample was used undigested in gels stained with Vistra Green (Amersham Biosciences) to determine DNA loading. Standard curves for the Southern blot were created by probing known quantities of the target fragment released from the plasmid construct in question. Standard curves for the loading control gel were created using purified DNA of the linearised plasmid construct.
Phototaxis assays
Qualitative phototaxis tests were performed as described in Darcy et al. (1994) and Fisher and Annesley (2006) by transferring a toothpick scraping of amoebae from a colony growing on a K. aerogenes lawn to the centre of charcoal agar plates (5% activated charcoal, 1.0% agar). Phototaxis was scored after a 48-h incubation at 21°C with a lateral light source. Slug trails were transferred to PVC discs, stained with Coomassie Blue and digitised. The orientation of the slug migration was analysed using directional statistics (Fisher et al. 1981; Fisher and Annesley 2006).
Growth in liquid media
Dictyostelium wild type and transformant cell lines were grown axenically in HL-5 with no antibiotics to exponential phase, subcultured into fresh HL-5 medium (no antibiotics) at an initial cell density of 1 × 104 cells/ml and counted using a haemocytometer at 8–12 h intervals over a period of 100 h. The cell densities were then analysed by log-linear regression using the R programming environment computer software to determine the generation time during the exponential phase of growth.
Western blot and immunoprecipitation
For Western blotting, 1 × 107 cells were lysed in 200 μL of lysis buffer (50 mM tris–HCl, 150 mM NaCl and 0.02% Triton-X100), centrifuged (10,000×g for 20 min) and the supernatant transferred to a new tube. A 15 μL aliquot of this crude protein extract in 4× loading buffer (62.5 mM tris–HCl, pH 6.8; 25% glycerol; 2% SDS; 0.01% bromophenol blue) was boiled for 5 min and loaded onto a 10% polyacrylamide gel for SDS–PAGE. The separated proteins were transferred to nitrocellulose membrane (Bio-Rad) and incubated in primary rabbit anti-NdkC (affinity purified, a kind gift from Dr. M.L. Lacombe, 1:1000), rabbit anti-AMPKα (Bokko et al. 2007) (1:500), rabbit anti-pThr-AMPK (a kind gift from Dr. D. Stapleton) (1:1,000) or mouse anti-actin (Santa Cruz Biotech, 1:1,000) antibodies followed by incubation in secondary anti-rabbit (Santa Cruz Biotech.) or anti-mouse (Amersham) antibodies, both at 1:2,500. Coimmunoprecipitations were performed using 1 × 107 cells in 300 μL lysis buffer and FLAG M2 affinity gel (Sigma) according to the manufacturer's instructions.
Immunofluorescence
Exponentially growing AX2 cells were incubated for 1 h in HL-5 followed by incubation for 45 min in LoFlo HL-5 (control) or in 3 × diluted LoFlo HL-5 media for 45 min, presenting hypotonic conditions. The cells (1 × 107 cells/mL) were starved for 7 h at 22°C in development buffer (5 mM Na2HPO4, 5 mM KH2PO4, 1 mM CaCl2, 2 mM MgCl2) on an orbital shaker (150 rpm). AX2 cells prepared as above were seeded onto glass coverslips in 6-well Costar plate chambers, allowed to settle and attach for 30 min at 22°C, fixed in 4% paraformaldehyde, permeabilised in chilled methanol for 3 min, blocked (1% BSA, 1% gelatin, 0.05% TritonX-100) for 30 min and labelled with primary antibodies (anti-NdkC and anti-actin) overnight at 4°C. Next day, the cells were incubated in anti-mouse Alexa 594 and anti-rabbit Alexa 488 secondary antibodies for 1 h at room temperature. Finally, the cells were overlaid with 4′, 6-diamino-2-phenyl-indol (DAPI, Sigma) for 5 min and mounted in 90% glycerol in PBS. Cells were observed on an Olympus BX61TRF microscope, and digital images were captured using an Olympus U-CMAD3 camera.
Results
Genetic interaction between AMPK and NDPK
Creation of transformants
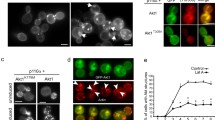
To investigate a genetic interaction between NDPK and AMPK and to investigate whether NDPK plays a role in the pathway leading to mitochondrial disease pathogenesis, NDPK expression was knocked down either in mitochondrially diseased cells or in strains overexpressing a truncated, constitutively active form of AMPKα, AMPKαT (Bokko et al. 2007). Mitochondrially diseased cells were created by antisense-inhibiting expression of chaperonin 60, a molecular chaperone which is responsible for the correct folding of mitochondrial proteins. Stable cell lines were created by transforming AX2 wild-type cells with the relevant constructs or pairs of constructs for antisense inhibition of NDPK and Cpn60 or overexpression of AMPKαT. Multiple, independent stable transformants were isolated and retained for each construct or combination of constructs. The integration of such constructs into random, independent sites in the genome is accompanied by rolling circle replication of the plasmid, so that each transformant contains a different number of copies of each construct used, ranging typically from a few to several hundred (Barth et al. 1998). The expression levels of the gene in question are correlated with the number of copies of the relevant construct (Bokko et al. 2007; Kotsifas et al. 2002) as determined by quantitative Southern blotting. Control transformants containing the AMPKαT overexpression and Cpn60 antisense-inhibition constructs with the previously reported copy numbers were also used (Bokko et al. 2007; Kotsifas et al. 2002). Western analysis using a pThr-AMPK antibody of strains overexpressing AMPKαT showed that the AMPKαT is phosphorylated and active (Fig. 1). The blot also shows that AMPKα is phosphorylated in strains overexpressing full-length AMPKα.
AMPKα is phosphorylated and active in strains overexpressing AMPKα or AMPKαT. Western blot analysis of wild-type cells (lane 1), two individual transformants overexpressing full-length AMPKα (lanes 2 and 3) and two individual transformants overexpressing AMPKαT (lanes 4 and 5) are shown. The blot was detected using a pThr-AMPKα specific antibody and clearly shows that the endogeneous AMPKα is phosphorylated at low levels in all strains, that there are higher levels of phosphorylated AMPKα in the strains overexpressing the full-length protein and that the overexpressed truncated form, AMPKαT, is heavily phosphorylated
NDPK knock down can rescue the morphological and phototactic defects in mitochondrial disease
The Dictyostelium life cycle permits analysis of both the directional migration of the motile multicellular slug form and the gross morphology of the multicellular fruiting body. Mitochondrial disease and chronic activation of AMPK are known to cause defects in fruiting body morphology and slug phototaxis (Bokko et al. 2007; Kotsifas et al. 2002). Cotransformants were studied phenotypically to determine the effect of NDPK downregulation in mitochondrial diseased cells and cells overexpressing AMPKαT. Phototaxis assays and fruiting body morphology analysis were performed and compared to wild type and Cpn60 and AMPKαT transformants. As evident in Figs. 2 and 3, knock down of NDPK in these strains is sufficient to rescue the phototaxis and morphological defects, respectively. To our knowledge, this is the first genetic evidence that these enzymes, which reportedly regulate a number of the same cellular functions, do in fact interact functionally in vivo.
NDPK knock down rescues the phototaxis defect caused by chronic AMPK signalling. The slug trails were plotted and digitised with the light source to the right of the figure. The single transformants containing either chaperonin 60 antisense inhibition constructs or AMPKαT overexpression constructs show low accuracies of phototaxis. The cotransformants in which NDPK had been downregulated through antisense inhibition show wild-type phototaxis with high accuracies of phototaxis. Copy numbers are written in brackets above each transformant and are indicative of expression levels
NDPK knock down suppresses the fruiting body morphology defect caused by chronic AMPK signalling. Transformants in which AMPKαT had been overexpressed or chaperonin 60 had been antisense inhibited show altered fruiting body morphology with shorter, thicker stalks. The cotransformants in the right of the figure resemble wild-type fruiting bodies with long, slender stalks. The copy number of each construct is listed above each fruiting body and is indicative of expression levels of those proteins
NDPK knock down can rescue the growth defects caused by AMPK signalling
In the unicellular, vegetative phase of its life cycle, laboratory strains of Dictyostelium are able to grow axenically as amoebae in liquid medium which they take up by macropinocytosis to fuel metabolism. The growth in axenic medium is inhibited in cells in which AMPKαT is overexpressed or Cpn60 is underexpressed (by antisense inhibition, thereby compromising mitochondrial protein folding and ATP generation) (Bokko et al. 2007; Kotsifas et al. 2002). In both cases, the resulting phenotypes are mediated by chronic AMPK hyperactivity. When NDPK expression was knocked down in cotransformants that either overexpressed AMPKαT or underexpressed Cpn60, the growth in axenic medium (Fig. 4) was restored to wild-type levels. These results indicate that NDPK is required for AMPK-mediated inhibition of growth.
Antisense inhibition of NDPK expression suppresses the axenic growth defect of mitochondrially diseased cells and AMPKαΤ overexpressing cells. The growth rates in axenic medium for cotransformants containing chaperonin 60 antisense constructs and NDPK antisense constructs and transformants containing chaperonin 60 antisense constructs alone are in (a). Growth rates of cotransformants containing AMPKαT overexpression and NDPK antisense constructs or AMPKαT overexpression alone are in (b). In the case of the Cpn60 antisense and the AMPKαT overexpression strains, data from Bokko et al. (2007) were normalized against the AX2 control, and the error bars are 95% confidence limits determined from the growth curve regression analysis. In the case of the strains containing the NDPK antisense constructs, data were normalized against the mean of all the NDPK antisense strains and the AX2 control. The error bars for these strains are standard errors of the mean from two independent experiments. Multiple log-linear regression analysis confirmed that the intercepts of the regression lines were not significantly different from 100% (P > 0.1) and that the slopes of the lines relating generation time to the copy numbers of the Cpn60 antisense (a) or AMPKαT (b) constructs were not significantly different from zero (P > 0.1) in cells also containing the NDPK antisense construct. However, in transformants containing either the Cpn60 antisense (a) or AMPKαT (b) construct alone, the slopes of the lines were highly significant and different from those for transformants also containing the NDPK antisense construct (P < 0.01). Regression lines were fitted to an exponential function (which would appear as a straight line on a log-linear plot). In the case of the cotransformants, the regression analysis was conducted as a separate function of NDPK antisense copy number (a, b) or the Cpn60 antisense copy number (a only) or the AMPKαT copy number (b only). However, since the slope of the line for cotransformants did not differ significantly from zero in each case, only one line is plotted for clarity (the NDPK antisense copy number regression in (a) and the AMPKαT regression in (b))
NDPK does not interact biochemically with AMPK
Having observed a genetic interaction between NDPK and AMPK, we wanted to see if the two proteins interact biochemically and thus physically associate with one another in the cell. To detect such physical interactions, we performed coimmunoprecipitation experiments. Cells overexpressing FLAG-tagged NDPK in combination with either hexahistidine-tagged AMPKαT or the full-length AMPKα subunit were used. Coimmunoprecipitations using crude protein extracts from these cells failed to reveal a physical interaction between the two proteins (Fig. 5). Additional experiments were performed to increase the activity of AMPK in these cells by growing the cells in hypotonic solution or starving the cells in development buffer, but in all cases, the results were the same (not shown).
Coimmunoprecipitation shows no biochemical interaction between AMPK and NDPK. Coimmunoprecipitations were performed using FLAG beads in the pull down and immunodetection with the antibodies listed to the right of the figure. Proteins which are pulled down by the FLAG beads are present in the pellet lanes, and those that are not are present in the supernatant lanes. The strains used are indicated at the top of each lane, AX2 wild-type strain, two independent transformants expressing full-length AMPK and two independent transformants expressing AMPKαT. The AMPKα and AMPKαT overexpressing strains also coexpress FLAG-NDPK. FLAG-NDPK (present at a higher density than NDPK alone) and NDPK were both pulled down from cells expressing FLAG-NDPK but not from the AX2 parental control. Endogenous NDPK can be seen in the supernatant in the AX2 lanes as it is not pulled down by the FLAG antibody. Neither the hexahistidine-tagged, full-length AMPKα subunit nor the truncated AMPKαT was present in the pellet, but both remained in the supernatant. β-actin was used as a negative control, was not present in any of the pellet lanes and remained in the supernatant. Coimmunoprecipitations were performed multiple times, and representative results are shown
Discussion
It has become increasingly clear that NDPK has many functions apart from its housekeeping role as a phosphotransfer protein. Many of these roles have been shown to result from interactions of NDPK with other proteins. For example, NDPK has been shown to play a role in endocytosis and cell adhesion, and this is facilitated by various interactions including the interaction of NDPK-A with ARF6-GTP. This protein is involved in the disassembly of adherens junctions, a premetastatic event in cancer. ARF6-GTP interacts with and recruits NDPK-A to the cell membrane which aids in the process of endocytosis (Palacios et al. 2002). Additionally, NDPK-B has been shown to directly interact with integrin cytoplasmic domain-associated protein (ICAP-1α) and forms a complex with ICAP-1α, β-integrin and Rac1 on the cell membrane during early adhesion (Fournier et al. 2003, 2002). NDPK also plays a role in vesicle trafficking as was shown by Krishnan et al. (2001) in Drosophila where NDPK regulates dynamin-dependent internalisation processes such as synaptic vesicle endocytosis and vesicle-dependent turnover of receptors. Another interaction that has been reported is an interaction with the cellular energy sensor AMPK, but this remains controversial, having not yet been confirmed for other cell types.
AMP-activated kinase is activated by a rise in cellular AMP concentration which occurs in mitochondrial disease or in other stress conditions. Once activated, AMPK phosphorylates multiple downstream substrates to switch off energy-consuming pathways and activate energy-producing ones. In Dictyostelium, chronic activation of AMPK leads to defects in several energy-consuming pathways such as growth in axenic medium as single cells, phototaxis (oriented movement towards light) in the multicellular migratory slug stage and formation of the multicellular fruiting body. Our results have shown that NDPK is required in this process as downregulation of NDPK in cells with chronically activated AMPK was able to rescue all the phenotypic defects. These findings support the work by Treharne et al. (2009) showing that NDPK interacts with AMPK in mammalian epithelial cell membranes.
Our coimmunoprecipitation experiments however suggested that the genetic interaction observed in Dictyostelium cells is not mediated by a direct interaction between AMPK and NDPK. If NDPK was channelling substrates as observed in epithelial membranes, then a close association would be expected. In Dictyostelium, this does not appear to be the case, suggesting that the genetic interaction we observed is indirect, perhaps through a common pathway such as the TOR pathways which control growth, cytoskeletal movement, etc. In the mammalian example, the authors reported a tight interaction between NDPK and the cystic fibrosis protein, CFTR (Treharne et al. 2009). They also found that, when AMPK and NDPK were added together in vitro, the net phosphorylation of NDPK was AMP dependent. One possibility suggested by Treharne et al. (2009) is that NDPK locally channels ATP produced from GTP plus an ADP to AMPK in order for it to phosphorylate its downstream targets.
Given that an interaction between AMPK and CFTR is also known to occur (Hallows et al. 2000), it is possible that CFTR acts specifically in epithelial cells as a scaffold to colocalize and facilitate a direct substrate channelling interaction between NDPK and AMPK. However, there are several possible alternative explanations for our inability to detect an AMPK–NDPK interaction by coimmunoprecipitation. Firstly, the pull-down experiments involved epitope-tagged recombinant proteins, and the tags may have interfered with the interaction. Nonrecombinant proteins could not be used because of the lack of antiNDPK and antiAMPK antibodies suitable for immunoprecipitation. NDPK forms hexameric structures in vivo, and the FLAG tag on NDPK apparently did not prevent this interaction between NDPK subunits since both tagged and untagged NDPK subunits were immunoprecipitated with the antiFLAG antibody. Nevertheless, heterologous interactions with other proteins such as AMPK may have been adversely affected. A second possibility is that NDPK and AMPK interact only transiently and/or that their interaction is unstable and disrupted during isolation of the proteins. Finally, the coimmunoprecipitation may have pulled down NDPK and AMPK from the wrong subcellular pools. The NDPK studied here resides in the cytoplasm but is enriched at the cell cortex (unpublished immunofluorescence data) as in mammalian cells. The mammalian work which shows a direct interaction between NDPK and AMPK involved the use of cell membrane extracts and not cytosolic extracts. This is consistent with there being at least two pools of NDPK, one of which is cytosolic while the other is membrane bound. Mammalian AMPK and NDPK interact at the membrane, and their colocalisation disappears under hypoxic conditions (Treharne et al. 2009). Thus, the two locations may reflect different functions of NDPK and possibly different protein interactions under different conditions.
Although we could not detect a direct biochemical interaction between NDPK and AMPK, our data provide the first unequivocal evidence for a functional genetic interaction between these two proteins that jointly regulate a variety of cellular functions and play dramatically pleiotropic roles in healthy and diseased cells.
References
Annesley SJ, Fisher PR (2009) Dictyostelium discoideum–a model for many reasons. Mol Cell Biochem 329:73–91
Arnaud-Dabernat S, Bourbon PM, Dierich A, Le Meur M, Daniel J-Y (2003) Knockout mice as model systems for studying nm23/NDP kinase gene functions. J Bioenerg Biomembr 35:19–30
Arnaud-Dabernat S, Masse K, Smani M, Peuchant E, Landry M, Bourbon P-M, Le Floch R, Daniel J-Y, Larou M (2004) Nm23-M2/NDP kinase B induces endogenous c-myc and nm23-M1/NDP kinase A overexpression in BAF3 cells. Both NDP kinases protect the cells from oxidative stress-induced death. Exp Cell Res 301:293–304
Barth C, Fraser DJ, Fisher PR (1998) Co-insertional replication is responsible for tandem multimer formation during plasmid integration into the Dictyostelium genome. Plasmid 39:141–153
Biggs J, Tripoulas N, Hersperger E, Dearolf C, Shearn A (1988) Analysis of the lethal interaction between the prune and Killer of prune mutations of Drosophila. Genes Develop 2:1333–1343
Boissan M, Dabernat S, Peuchant E, Schlattner U, Lascu I, Lacombe M-L (2009) The mammalian Nm23/NDPK family: from metastasis control to cilia movement. Mol Cell Biochem 329:51–62
Bokko PB, Francione L, Ahmed AU, Annesley SJ, Huang X, Khurana T, Kimmel AR, Fisher PR (2007) Diverse mitochondrial cytopathologies are caused by AMPK signalling. Mol Biol Cell 18:1874–1886
Bosnar MH, De Gunzburg J, Bago R, Brecevic L, Weber I, Pavelic J (2004) Subcellular localisation of A and B Nm23/NDPK subunits. Exp Cell Res 298:275–284
Chang CL, Zhu XX, Thoraval DH, Ungar D, Rawwas J, Hora N, Strahler JR, Hanash SM, Radany E (1994) Nm23-H1 mutation in neuroblastoma. Nature 370:335–336
Cherflis J, Morera S, Lascu I, Lacombe ML, Veron M, Janin J (1994) X-ray structure of nucleoside diphosphate kinase complexed with thymidine diphosphate and Mg2+ at 2-Å resolution. Biochem 3:9062–9069
Darcy PK, Wilczynska Z, Fisher PR (1994) Genetic analysis of Dictyostelium slug phototaxis mutants. Genetics 137:977–985
Dumas C, Lascu I, Morera S, Glaser P, Fourme R, Wallet V, Lacombe ML, Veron M (1992) X-ray structure of nucleoside diphosphate kinase. EMBO J 11:3203–3208
Eichinger L, Pachebat JA, Glockner G, Rajandream MA, Sucgang R, Berriman M, Song J, Olsen R, Szafranski K et al (2005) The genome of the social amoeba Dictyostelium discoideum. Nature 435:43–57
Fisher PR, Annesley SJ (2006) Slug phototaxis, thermotaxis and spontaneous turning behaviour. In: Eichinger L and Rivero F (eds) Dictyostelium discoideum protocols. Methods in molecular biology. Humana Press 346:137–170
Fisher PR, Smith E, Williams KL (1981) An extracellular chemical signal controlling phototactic behaviour by D. discoideum slugs. Cell 23:799–807
Fournier HN, Dupe-Manet S, Bouvard D, Lacombe ML, Marie C, Block MR, Albiges-Rizo C (2002) Integrin cytoplasmic domain-associated protein 1 alpha (ICAP-1 alpha) interacts directly with the metastasis suppressor nm23-H2, and both proteins are targeted to newly formed cell adhesion sites upon integrin engagement. J Biol Chem 277:20895–20902
Fournier H-N, Albiges-Rizo C, Block MR (2003) New insights into Nm23 control of cell adhesion and migration. J Bioenerg Biomembr 35:81–87
Hallows KR, Raghuram V, Kemp BE, Witters LA, Foskett JK (2000) Inhibition of cystic fibrosis transmembrane conductance regulator by novel interaction with the metabolic sensor AMP-activated protein kinase. J Clin Invest 105:1711–1721
Hardie G (1992) Regulation of fatty acid and cholesterol metabolism by the AMP-activated protein kinase. Biochim Biophys Acta 1123(3):231–238
Karlsson A, Mesnildrey S, Xu Y, Morera S, Janin J, Veron M (1996) Nucleoside diphosphate kinase. Investigation of the intersubunit contacts by site-directed mutagenesis and crystallography. J Biol Chem 271:19928–19934
Kotsifas M, Barth C, Lay ST, de Lozanne A, Fisher PR (2002) Chaperonin 60 and mitochondrial disease in Dictyostelium. J Muscle Res Cell Motil 23:839–852
Krishnan KS, Rikhy R, Rao S, Shivalkar M, Mosko M, Narayanan R, Etter P, Estes PS, Ramaswami M (2001) Nucleoside diphosphate kinase, a source of GTP, is required for dynamin-dependent synaptic vesicle recycling. Neuron 30:197–210
Lacombe M-L, Wallet V, Troll H, Veron M (1990) Functional cloning of a nucleoside diphosphate kinase from Dictyostelium discoideum. J Biol Chem 265:10012–10018
Lacombe M-L, Milon L, Munier A, Mehus JG, Lambeth DO (2000) The human Nm23/nucleoside diphosphate kinase. J Bioenerg Biomembr 32:247–258
Morera S, Lascu I, Dumas C, LeBras G, Briozzo P, Veron M, Janin J (1994a) Adenosine 5′-diphosphoate binding and the active site of nucleoside diphosphate kinase. Biochem 33:459–467
Morera S, LeBras G, Lascu I, Lacombe ML, Veron M, Janin J (1994b) Refined X-ray structure of Dictyostelium discoideum nucleoside diphosphate kinase at 1.8Å resolution. J Mol Biol 243:873–890
Morera S, Chiadmi M, LeBras G, Lascu I, Janin J (1995) Mechanism of phosphate transfer by nucleoside diphosphate kinase: x-ray structures of the phosphohistidine intermediate of the enzymes from Drosophila and Dictyostelium. Biochem 34:11062–11070
Muimo R, Crawford RM, Mehta A (2006) Nucleoside diphosphate kinase A as a controller of AMP-kinase in airway epithelia. J Bioenerg Biomembr 38:181–187
Nellen W, Slamin C, Firtel RA (1984) DNA-mediated transformation in Dictyostelium discoideum: regulated expression of an actin gene fusion. Mol Cell Biol 4:2890–2898
Onken B, Driscoll M (2010) Metformin induces a dietary restriction-like state and the oxidative stress response to extend C. elegans healthspan via AMPK, LKB1, and SKN-1. PLoS ONE 5:e8758
Palacios F, Schweitzer JK, Boshans RL, D’Souza-Schorey C (2002) ARF6-GTP recruits Nm23-H1 to facilitate dynamin-mediated endocytosis during adherens junctions disassembly. Nat Cell Biol 4:929–936
Postel EH, Wohlman I, Zou X, Juan T, Sun N, D'Agostin D, Cuellar M, Choi T, Notterman DA, La Perle KMD (2009) Targeted deletion of Nm23/nucleoside diphosphate kinase A and B reveals their requirement for definitive erythropoieses in the mouse embryo. Dev Dyn 238:775–787
Sussman M (1966) Biochemical and genetic methods in the study of cellular slime mold development. Methods Cell Physiol 2:397–410
Treharne KJ, Best OG, Mehta A (2009) The phosphorylation status of membrane-bound nucleoside diphosphate kinase in epithelia and the role of AMP. Mol Cell Biochem 329:107–114
Wagner PD, Vu N-D (1995) Phosphorylation of ATP-citrate lyase by nucleoside diphosphate kinase. J Biol Chem 270:21758–21764
Watts DJ, Ashworth JM (1970) Growth of myxameobae of the cellular slime mould Dictyostelium discoideum in axenic culture. Biochem J 119:171–174
Wilczynska Z, Fisher PR (1994) Analysis of a complex plasmid insertion in a phototaxis-deficient transformant of Dictyostelium discoideum selected on a Micrococcus luteus lawn. Plasmid 32:182–194
Wilczynska Z, Barth C, Fisher PR (1997) Mitochondrial mutations impair signal transduction in Dictyostelium discoideum slugs. Biochem Biophys Res Commun 234:39–43
Acknowledgements
This work was supported by grants to PRF from the Thyne Reid Memorial Trusts and the Australian Research Council and to AM from Wellcome Trust WT086370MA. Thanks to Dr. Lacombe for her kind gift of the NDPK antibody and Dr. D. Stapleton for his kind gift of the pThr-AMPK antibody.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License ( https://creativecommons.org/licenses/by-nc/2.0 ), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Annesley, S.J., Bago, R., Mehta, A. et al. A genetic interaction between NDPK and AMPK in Dictyostelium discoideum that affects motility, growth and development. Naunyn-Schmiedeberg's Arch Pharmacol 384, 341–349 (2011). https://doi.org/10.1007/s00210-011-0615-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00210-011-0615-0