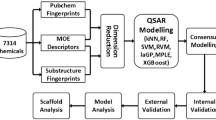
Abstract
Organophosphates (OPs) are hazardous chemicals widely used in industry and agriculture. Distribution of their residues in nature causes serious risks to humans, animals, and plants. To reduce hazards from OPs, quantitative structure–activity relationship (QSAR) models for predicting their acute oral toxicity in rats and mice and inhibition constants concerning human acetylcholinesterase were developed according to the bioactivity data of 456 unique OPs. Based on robust, two-dimensional molecular descriptors and quantum chemical descriptors, which accurately reflect OP electronic structures and reactivities, the influences of eight machine-learning algorithms on the prediction performance of the QSAR models were explored, and consensus QSAR models were constructed. Several strict model validation indices and the results of applicability domain evaluations show that the established consensus QSAR models exhibit good robustness, practical prediction abilities, and wide application scopes. Poor correlation was observed between acute oral toxicity at the mammalian level and the inhibition constants at the molecular level, indicating that the acute toxicity of OPs cannot be evaluated only by the experimental data of enzyme inhibitory activity, their toxicokinetic characteristics must also be considered. The constructed QSAR models described herein provide rapid, theoretical assessment of the bioactivity of unstudied or unknown OPs, as well as guidance for making decisions regarding their regulation.






Similar content being viewed by others
Abbreviations
- OPs:
-
Organophosphates
- OECD:
-
Organization for Economic Co-operation and Development
- QSAR:
-
Quantitative structure activity relationship
- 2D:
-
Two dimensional
- QC:
-
Quantum chemical
- AD:
-
Application domain
- ML:
-
Machine learning
- AChE:
-
Acetylcholinesterase
- Ach:
-
Acetylcholine
- LD50 :
-
Median lethal dose
- hAChE:
-
Human acetylcholinesterase
- DFT:
-
Density functional theory
- MOE:
-
Molecular operating environment
- PLS:
-
Partial least square
- GB:
-
Gradient boosting
- RF:
-
Random forest
- XGBoost:
-
Extreme gradient boosting
- GP:
-
Gaussian process
- SVM:
-
Support vector machine
- RVM:
-
Relevance vector machine
- SMD:
-
Solvation model based on density
- NPA:
-
Natural population analysis
- MLR:
-
Multiple linear regression
- PCA:
-
Principal component analysis
- RMSE:
-
Root mean square error
- RFE-RF:
-
The recursive feature elimination based on random forest
References
Becke AD (1993) Density-functional thermochemistry. III. The role of exact exchange. J Chem Phys 98(7):5648–5653. https://doi.org/10.1063/1.464913
Bemis GW, Murcko MA (1996) The properties of known drugs. 1. Molecular frameworks. J Med Chem 39(15):2887–2893. https://doi.org/10.1021/jm9602928
Bermúdez-Saldaña JM, Cronin MTD (2006) Quantitative structure-activity relationships for the toxicity of organophosphorus and carbamate pesticides to the rainbow trout onchorhyncus mykiss. Pest Manag Sci 62(9):819–831. https://doi.org/10.1002/ps.1233
Bertero A, Chiari M, Vitale N et al (2020) Types of pesticides involved in domestic and wild animal poisoning in Italy. Sci Total Environ 707:136129. https://doi.org/10.1016/j.scitotenv.2019.136129
Bille L, Toson M, Mulatti P et al (2016) Epidemiology of animal poisoning: an overview on the features and spatio-temporal distribution of the phenomenon in the north-eastern Italian regions. Forensic Sci Int 266:440–448. https://doi.org/10.1016/j.forsciint.2016.07.002
Breiman L (2001) Random forests. Mach Learn 45(1):5–32. https://doi.org/10.1023/a:1010933404324
Caloni F, Cortinovis C, Rivolta M, Davanzo F (2016) Suspected poisoning of domestic animals by pesticides. Sci Total Environ 539:331–336. https://doi.org/10.1016/j.scitotenv.2015.09.005
Camacho-Mendoza RL, Aquino-Torres E, Cordero-Pensado V et al (2018) A new computational model for the prediction of toxicity of phosphonate derivatives using QSPR. Mol Divers 22(2):269–280. https://doi.org/10.1007/s11030-018-9819-2
Cao DS, Xu QS, Liang YZ, Chen X, Li HD (2010a) Prediction of aqueous solubility of druglike organic compounds using partial least squares, back-propagation network and support vector machine. J Chemom 24(9–10):584–595. https://doi.org/10.1002/cem.1321
Cao DS, Xu QS, Liang YZ, Zhang LX, Li HD (2010b) The boosting: a new idea of building models. Chemometr Intell Lab Syst 100(1):1–11. https://doi.org/10.1016/j.chemolab.2009.09.002
Cao DS, Liang YZ, Xu QS, Li HD, Chen X (2010c) A new strategy of outlier detection for QSAR/QSPR. J Comput Chem 31(3):592–602. https://doi.org/10.1002/jcc.21351
Cao DS, Liang YZ, Xu QS, Hu QN, Zhang LX, Fu GH (2011a) Exploring nonlinear relationships in chemical data using kernel-based methods. Chemometr Intell Lab Syst 107(1):106–115. https://doi.org/10.1016/j.chemolab.2011.02.004
Cao DS, Liang YZ, Xu QS, Zhang LX, Hu QN, Li HD (2011b) Feature importance sampling-based adaptive random forest as a useful tool to screen underlying lead compounds. J Chemom 25(4):201–207. https://doi.org/10.1002/cem.1375
Cao DS, Liu S, Fan L, Liang YZ (2014) QSAR analysis of the effects of OATP1B1 transporter by structurally diverse natural products using a particle swarm optimization-combined multiple linear regression approach. Chemometr Intell Lab Syst 130:84–90. https://doi.org/10.1016/j.chemolab.2013.10.011
Cao DS, Dong J, Wang NN et al (2015) In silico toxicity prediction of chemicals from EPA toxicity database by kernel fusion-based support vector machines. Chemometr Intell Lab Syst 146:494–502. https://doi.org/10.1016/j.chemolab.2015.07.009
Chandrashekar G, Sahin F (2014) A survey on feature selection methods. Comput Electr Eng 40(1):16–28. https://doi.org/10.1016/j.compeleceng.2013.11.024
Chattaraj PK, Giri S, Duley S (2011) Update 2 of: electrophilicity index. Chem Rev 111(2):PR43–PR75. https://doi.org/10.1021/cr100149p
Chen BY (2011) Hydrolytic stabilities of halogenated disinfection byproducts: review and rate constant quantitative structure-property relationship analysis. Environ Eng Sci 28(6):385–394. https://doi.org/10.1089/ees.2010.0196
Chen TQ, Guestrin C (2016) XGBoost: a scalable tree boosting system. Assoc Comp Mach N Y. https://doi.org/10.1145/2939672.2939785
Chen BY, Zhang T, Bond T, Gan YQ (2015) Development of quantitative structure activity relationship (QSAR) model for disinfection byproduct (DBP) research: a review of methods and resources. J Hazard Mater 299:260–279. https://doi.org/10.1016/j.jhazmat.2015.06.054
Cherkasov A, Muratov EN, Fourches D et al (2014) QSAR modeling: where have you been? where are you going to? J Med Chem 57(12):4977–5010. https://doi.org/10.1021/jm4004285
Chirico N, Gramatica P (2012) Real external predictivity of QSAR models. Part 2. New intercomparable thresholds for different validation criteria and the need for scatter plot inspection. J Chem Inf Model 52(8):2044–2058. https://doi.org/10.1021/ci300084j
Čolović MB, Krstić DZ, Lazarević-Pašti TD, Bondžić AM, Vasić VM (2013) Acetylcholinesterase inhibitors: pharmacology and toxicology. Curr Neuropharmacol 11(3):315–335. https://doi.org/10.2174/1570159x11311030006
Cortes C, Vapnik V (1995) Support vector networks. Mach Learn 20(3):273–297. https://doi.org/10.1007/bf00994018
Dennington R, Keith TA, Millam JM (2016) GaussView, 6th edn. Semichem Inc., Shawnee Mission
Devillers J (2004) Prediction of mammalian toxicity of organophosphorus pesticides from QSTR modeling. SAR QSAR Environ Res 15(5–6):501–510. https://doi.org/10.1080/10629360412331297443
Ding XQ, Ding JJ, Li DY, Pan L, Pei CX (2018) Toxicity prediction of organophosphorus chemical reactivity compounds based on conceptual DFT. Acta Phys Chim Sin 34(3):314–322. https://doi.org/10.3866/pku.whxb201709042
dos Santos VMR, Donnici CL, DaCosta JBN, Caixeiro JMR (2007) Organophosphorus pentavalent compounds: history, synthetic methods of preparation and application as insecticides and antitumor agents. Quim Nova 30(1):159–170. https://doi.org/10.1590/s0100-40422007000100028
Eriksson L, Jaworska J, Worth AP, Cronin MTD, McDowell RM, Gramatica P (2003) Methods for reliability and uncertainty assessment and for applicability evaluations of classification- and regression-based QSARs. Environ Health Perspect 111(10):1361–1375. https://doi.org/10.1289/ehp.5758
Fearn T (2013) Gaussian process regression. NIR news 24(6):23–24. https://doi.org/10.1255/nirn.1392
Fourches D, Muratov E, Tropsha A (2010) Trust, but verify: on the importance of chemical structure curation in cheminformatics and QSAR modeling research. J Chem Inf Model 50(7):1189–1204. https://doi.org/10.1021/ci100176x
Franjesevic AJ, Sillart SB, Beck JM, Vyas S, Callam CS, Hadad CM (2019) Resurrection and reactivation of acetylcholinesterase and butyrylcholinesterase. Chem Eur J 25(21):5337–5371. https://doi.org/10.1002/chem.201805075
Friedman JH (2002) Stochastic gradient boosting. Comput Stat Data Anal 38(4):367–378. https://doi.org/10.1016/s0167-9473(01)00065-2
Frisch MJ, Trucks GW, Schlegel HB et al (2016) Gaussian 16 Rev. B.01. Gaussian Inc, Wallingford
Fu L, Liu L, Yang ZJ et al (2020) Systematic modeling of log D7.4 based on ensemble machine learning, group contribution, and matched molecular pair analysis. J Chem Inf Model 60(1):63–76. https://doi.org/10.1021/acs.jcim.9b00718
Fujiwara S, Yamashita F, Hashida M (2002) Prediction of Caco-2 cell permeability using a combination of MO-calculation and neural network. Int J Pharm 237(1–2):95–105. https://doi.org/10.1016/s0378-5173(02)00045-5
Gadaleta D, Vukovic K, Toma C et al (2019) SAR and QSAR modeling of a large collection of LD50 rat acute oral toxicity data. J Cheminform 11(1):58. https://doi.org/10.1186/s13321-019-0383-2
García-Domenech R, Alarcón-Elbal P, Bolas G et al (2007) Prediction of acute toxicity of organophosphorus pesticides using topological indices. SAR QSAR Environ Res 18(7–8):745–755. https://doi.org/10.1080/10629360701698712
Golbraikh A, Tropsha A (2002) Beware of q2! J Mol Graph Model 20(4):269–276. https://doi.org/10.1016/s1093-3263(01)00123-1
Golbraikh A, Shen M, Xiao ZY, Xiao YD, Lee KH, Tropsha A (2003) Rational selection of training and test sets for the development of validated QSAR models. J Comput Aided Mol Des 17(2):241–253. https://doi.org/10.1023/a:1025386326946
Gramatica P, Sangion A (2016) A historical excursus on the statistical validation parameters for QSAR models: a clarification concerning metrics and terminology. J Chem Inf Model 56(6):1127–1131. https://doi.org/10.1021/acs.jcim.6b00088
Grigoryan H, Schopfer LM, Peeples ES et al (2009) Mass spectrometry identifies multiple organophosphorylated sites on tubulin. Toxicol Appl Pharmacol 240(2):149–158. https://doi.org/10.1016/j.taap.2009.07.020
Gupta PK (2004) Pesticide exposure—Indian scene. Toxicology 198(1–3):83–90. https://doi.org/10.1016/j.tox.2004.01.021
Hamadache M, Benkortbi O, Hanini S, Amrane A, Khaouane L, Moussa CS (2016) A quantitative structure activity relationship for acute oral toxicity of pesticides on rats: validation, domain of application and prediction. J Hazard Mater 303:28–40. https://doi.org/10.1016/j.jhazmat.2015.09.021
Hansch C, Maloney PP, Fujita T, Muir RM (1962) Correlation of biological activity of phenoxyacetic acids with Hammett substituent constants and partition coefficients. Nature 194(4824):178–180. https://doi.org/10.1038/194178b0
Helland IS (2001) Some theoretical aspects of partial least squares regression. Chemometr Intell Lab Syst 58(2):97–107. https://doi.org/10.1016/s0169-7439(01)00154-x
Huang WK, Geng L, Deng R et al (2015) Prediction of human clearance based on animal data and molecular properties. Chem Biol Drug Des 86(5):990–997. https://doi.org/10.1111/cbdd.12567
Johnson H, Kenley RA, Rynard C, Golub MA (1985) QSAR for cholinesterase inhibition by organophosphorus esters and CNDO/2 calculations for organophosphorus ester hydrolysis. Quant Struct Act Relat 4(4):172–180. https://doi.org/10.1002/qsar.19850040406
Kar S, Sanderson H, Roy K, Benfenati E, Leszczynski J (2020) Ecotoxicological assessment of pharmaceuticals and personal care products using predictive toxicology approaches. Green Chem 22(5):1458–1516. https://doi.org/10.1039/c9gc03265g
Kennard RW, Stone LA (1969) Computer aided design of experiments. Technometrics 11(1):137–148. https://doi.org/10.1080/00401706.1969.10490666
Khan K, Roy K, Benfenati E (2019) Ecotoxicological QSAR modeling of endocrine disruptor chemicals. J Hazard Mater 369:707–718. https://doi.org/10.1016/j.jhazmat.2019.02.019
Krewski D, Acosta D Jr, Andersen M et al (2010) Toxicity testing in the 21st century: a vision and a strategy. J Toxicol Env Heal B 13(2–4):51–138. https://doi.org/10.1080/10937404.2010.483176
Lee S, Barron MG (2016) A mechanism-based 3D-QSAR approach for classification and prediction of acetylcholinesterase inhibitory potency of organophosphate and carbamate analogs. J Comput Aided Mol Des 30(4):347–363. https://doi.org/10.1007/s10822-016-9910-7
Liu SB (2009) Conceptual density functional theory and some recent developments. Acta Phys Chim Sin 25(3):590–600. https://doi.org/10.3866/pku.whxb20090332
Liu Z, Lu T, Chen Q (2021) Intermolecular interaction characteristics of the all-carboatomic ring, cyclo[18]carbon: focusing on molecular adsorption and stacking. Carbon 171:514–523. https://doi.org/10.1016/j.carbon.2020.09.048
Lu T, Chen FW (2012) Multiwfn: a multifunctional wavefunction analyzer. J Comput Chem 33(5):580–592. https://doi.org/10.1002/jcc.22885
Ma YX, Xie ZY, Lohmann R, Mi WY, Gao GP (2017) Organophosphate ester flame retardants and plasticizers in ocean sediments from the north pacific to the arctic ocean. Environ Sci Technol 51(7):3809–3815. https://doi.org/10.1021/acs.est.7b00755
Marenich AV, Cramer CJ, Truhlar DG (2009) Universal solvation model based on solute electron density and on a continuum model of the solvent defined by the bulk dielectric constant and atomic surface tensions. J Phys Chem B 113(18):6378–6396. https://doi.org/10.1021/jp810292n
Minasny B, McBratney AB (2008) Regression rules as a tool for predicting soil properties from infrared reflectance spectroscopy. Chemometr Intell Lab Syst 94(1):72–79. https://doi.org/10.1016/j.chemolab.2008.06.003
Nepovimova E, Kuca K (2018) Chemical warfare agent Novichok—mini-review of available data. Food Chem Toxicol 121:343–350. https://doi.org/10.1016/j.fct.2018.09.015
Netzeva TI, Worth AP, Aldenberg T et al (2005) Current status of methods for defining the applicability domain of (quantitative) structure-activity relationships—the report and recommendations of ECVAM workshop 521,2. ATLA-Altern Lab Anim 33(2):155–173. https://doi.org/10.1177/026119290503300209
Obrezanova O, Csanyi G, Gola JMR, Segall MD (2007) Gaussian processes: a method for automatic QSAR modeling of ADME properties. J Chem Inf Model 47(5):1847–1857. https://doi.org/10.1021/ci7000633
Palm K, Luthman K, Ungell AL, Strandlund G, Artursson P (1996) Correlation of drug absorption with molecular surface properties. J Pharm Sci 85(1):32–39. https://doi.org/10.1021/js950285r
Pesticide Action Network International, 2021. Pan international consolidated list of banned pesticides http://pan-international.org/pan-international-consolidated-list-of-banned-pesticides. Accessed 10 Jan 2021
Plyamovatyi AK, Vandyukova II, Shagidullin RR, Makhaeva GF, Malygin VV, Gorbunov SM (1997) Study of the relationship between spatial structure and anticholinesterase activity of o-phosphorylate oximes. Pharm Chem J 31(4):199–204. https://doi.org/10.1007/bf02464156
PubChem (2021) https://pubchem.ncbi.nlm.nih.gov Accessed 10 Jan 2021
Raies AB, Bajic VB (2016) In silico toxicology: computational methods for the prediction of chemical toxicity. WIRES Comput Mol Sci 6(2):147–172. https://doi.org/10.1002/wcms.1240
RDKit (2021) http://www.rdkit.org. Accessed 10 Jan 2021
Roy PP, Roy K (2008) On some aspects of variable selection for partial least squares regression models. QSAR Comb Sci 27(3):302–313. https://doi.org/10.1002/qsar.200710043
Roy K, Das RN, Ambure P, Aher RB (2016) Be aware of error measures. Further studies on validation of predictive QSAR models. Chemometr Intell Lab Syst 152:18–33. https://doi.org/10.1016/j.chemolab.2016.01.008
Roy K, Ambure P, Kar S (2018) How precise are our quantitative structure-activity relationship derived predictions for new query chemicals? ACS Omega 3(9):11392–11406. https://doi.org/10.1021/acsomega.8b01647
Ruark CD, Hack CE, Robinson PJ, Anderson PE, Gearhart JM (2013) Quantitative structure-activity relationships for organophosphates binding to acetylcholinesterase. Arch Toxicol 87(2):281–289. https://doi.org/10.1007/s00204-012-0934-z
Rücker C, Rücker G, Meringer M (2007) y-Randomization and its variants in QSPR/QSAR. J Chem Inf Model 47(6):2345–2357. https://doi.org/10.1021/ci700157b
Schulz E, Speekenbrink M, Krause A (2018) A tutorial on Gaussian process regression: modelling, exploring, and exploiting functions. J Math Psychol 85:1–16. https://doi.org/10.1016/j.jmp.2018.03.001
Sheridan RP, Wang WM, Liaw A, Ma JS, Gifford EM (2016) Extreme gradient boosting as a method for quantitative structure-activity relationships. J Chem Inf Model 56(12):2353–2360. https://doi.org/10.1021/acs.jcim.6b00591
Shoombuatong W, Schaduangrat N, Nantasenamat C (2018) Towards understanding aromatase inhibitory activity via QSAR modeling. Excli J 17:688–708. https://doi.org/10.17179/excli2018-1417
Sidhu GK, Singh S, Kumar V, Dhanjal DS, Datta S, Singh J (2019) Toxicity, monitoring and biodegradation of organophosphate pesticides: a review. Crit Rev Env Sci Technol 49(13):1135–1187. https://doi.org/10.1080/10643389.2019.1565554
Storm JE, Rozman KK, Doull J (2000) Occupational exposure limits for 30 organophosphate pesticides based on inhibition of red blood cell acetylcholinesterase. Toxicology 150(1–3):1–29. https://doi.org/10.1016/s0300-483x(00)00219-5
Tannenbaum J, Bennett BT (2015) Russell and Burch’s 3Rs then and now: the need for clarity in definition and purpose. J Am Assoc Lab Anim 54(2):120–132
Tipping ME (2001) Sparse Bayesian learning and the relevance vector machine. J Mach Learn Res 1(3):211–244. https://doi.org/10.1162/15324430152748236
Tropsha A (2010) Best practices for QSAR model development, validation, and exploitation. Mol Inform 29(6–7):476–488. https://doi.org/10.1002/minf.201000061
Wang NN, Deng ZK, Huang C et al (2017) ADME properties evaluation in drug discovery: prediction of plasma protein binding using NSGA-II combining PLS and consensus modeling. Chemometr Intell Lab Syst 170:84–95. https://doi.org/10.1016/j.chemolab.2017.09.005
Wang LL, Ding JJ, Pan L et al (2021) Quantitative structure-toxicity relationship model for acute toxicity of organophosphates via multiple administration routes in rats and mice. J Hazard Mater 401:123724. https://doi.org/10.1016/j.jhazmat.2020.123724
Worek F, Thiermann H (2013) The value of novel oximes for treatment of poisoning by organophosphorus compounds. Pharmacol Therapeut 139(2):249–259. https://doi.org/10.1016/j.pharmthera.2013.04.009
Worek F, Wille T, Koller M, Thiermann H (2016) Toxicology of organophosphorus compounds in view of an increasing terrorist threat. Arch Toxicol 90(9):2131–2145. https://doi.org/10.1007/s00204-016-1772-1
Wu ZX, Lei TL, Shen C, Wang Z, Cao DS, Hou TJ (2019) ADMET evaluation in drug discovery. 19. Reliable prediction of human cytochrome P450 inhibition using artificial intelligence approaches. J Chem Inf Model 59(11):4587–4601. https://doi.org/10.1021/acs.jcim.9b00801
Xu Y, Johnson M (2002) Using molecular equivalence numbers to visually explore structural features that distinguish chemical libraries. J Chem Inf Comput Sci 42(4):912–926. https://doi.org/10.1021/ci025535l
Xu J, Wang L, Wang LX, Shen XL, Xu WL (2011) QSPR study of Setschenow constants of organic compounds using MLR, ANN, and SVM analyses. J Comput Chem 32(15):3241–3252. https://doi.org/10.1002/jcc.21907
Yang H, Du Z, Lv WJ, Zhang XY, Zhai HL (2019) In silico toxicity evaluation of dioxins using structure-activity relationship (SAR) and two-dimensional quantitative structure-activity relationship (2D-QSAR). Arch Toxicol 93(11):3207–3218. https://doi.org/10.1007/s00204-019-02580-w
Zhao J, Yu S (2013) Quantitative structure-activity relationship of organophosphate compounds based on molecular interaction fields descriptors. Environ Toxicol Pharmacol 35(2):228–234. https://doi.org/10.1016/j.etap.2012.11.018
Acknowledgements
This work was supported by the HKBU Strategic Development Fund project (SDF19-0402-P02). The studies meet with the approval of the university’s review board.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, L., Ding, J., Shi, P. et al. Ensemble machine learning to evaluate the in vivo acute oral toxicity and in vitro human acetylcholinesterase inhibitory activity of organophosphates. Arch Toxicol 95, 2443–2457 (2021). https://doi.org/10.1007/s00204-021-03056-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00204-021-03056-6