Abstract
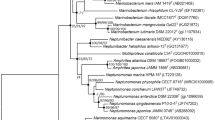
Three Gram-reaction-positive bacterial strains, designated KSW-18T, KSW2-22, and KSW4-11T, were isolated from seawater, and two dried seaweed samples collected at Gwakji Beach in Jeju, Republic of Korea, respectively, and their taxonomic positions were examined by a polyphasic approach. The 16S rRNA gene phylogeny showed that strain KSW4-11T was tightly associated with Microbacterium oleivorans NBRC 103075T, while strains KSW-18T and KSW2-22 formed a distinctive subline at the base of a clade including the above two strains. The three isolates showed high sequence similarity with one another (99.7–99.9%; 1–4 nt differences) and Microbacterium oleivorans (99.8–99.9%; 1–3 nt differences). The chemotaxonomic features were typical for the genus Microbacterium; Lysine as the diagnostic diamino acid and N-glycolylated muramic acid of the peptidoglycans, the predominant menaquinones of MK-11, MK-10 and MK-12, the major fatty acids of anteiso-C15:0 and anteiso-C17:0, and the major polar lipids including diphosphatidylglycerol, phosphatidylglycerol, and two or three unidentified glycolipids. In core genome-based phylogenetic tree, strains KSW-18T and KSW2-22 were closely associated with Microbacterium oleivorans NBRC 103075T, while strain KSW4-11T formed a distinctive subline at the base of a clade including the above three strains, in contrast to the 16S rRNA gene tree. Strains KSW-18T and KSW2-22 shared an OrthoANIu of 98.6% and a digital DNA–DNA hybridization of 87.6% with each other, representing that they were strains of a species, while the OrthoANIu and digital DNA–DNA hybridization values between strains KSW-18T and KSW4-11T, and between both of these isolates and all members of the genus Microbacterium were ≤86.5% and ≤30.7%, respectively. The analyses of overall genomic relatedness indices and phenotypic distinctness support that the three isolates represent two new species of the genus Microbacterium. Based on the results obtained here, Microbacterium aquilitoris sp. nov. (type strain KSW-18T = KCTC 49623T = NBRC 115222T) and Microbacterium gwkjiense sp. nov. (type strain KSW4-11T = KACC 23321T = DSM 116380T) are proposed.


Similar content being viewed by others
Data availability
The DDBJ/ENA/GenBank accession numbers for 16S rRNA gene and genome sequences are OR039658 and JAUZVV010000000, respectively.
Change history
04 April 2024
A Correction to this paper has been published: https://doi.org/10.1007/s00203-024-03929-1
References
Bellassi P, Fontana A, Callegari ML, Cappa F, Morelli L (2021) Microbacterium paulum sp. nov., isolated from microfiltered milk. Int J Syst Evol Microbiol 71:5119
Blin K, Shaw S, Steinke K, Villebro R, Ziemert N et al (2019) antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res 47:W81–W87
Dong K, Yang J, Lu S, Pu J, Lai X-H et al (2020) Microbacterium wangchenii sp. nov., isolated from faeces of Tibetan gazelles (Procapra picticaudata) on the Qinghai-Tibet Plateau. Int J Syst Evol Microbiol 70:1307–1314
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Fidalgo C, Riesco R, Henriques I, Trujillo ME, Alves A (2016) Microbacterium diaminobutyricum sp. nov., isolated from Halimione portulacoides, which contains diaminobutyric acid in its cell wall, and emended description of the genus Microbacterium. Int J Syst Evol Microbiol 66:4492–4500
Fitch WM (1971) Towards defining the course of evolution: minimum change for a specific tree topology. Syst Zool 20:406–416
Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P et al (2007) DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol 57:81–91
Jukes TH, Cantor CR (1969) Evolution of protein molecules. In: Munro HN (ed) Mammalian protein metabolism. Academic Press, New York, pp 21–132
Kim KK, Lee KC, Oh HM, Lee JS (2008) Microbacterium aquimaris sp. nov., isolated from seawater. Int J Syst Evol Microbiol 58:1616–1620
Kroppenstedt RM (1985) Fatty acid and menaquinone analysis of actinomycetes and related organisms. In: Goodfellow M, Minnikin DE (eds) Chemical methods in bacterial systematics. Academic Press, London, pp 173–199
Leão PN, Costa M, Ramos V, Pereira AR, Fernandes VC et al (2013) Antitumor activity of hierridin B, a cyanobacterial secondary metabolite found in both filamentous and unicellular marine strains. PLoS ONE 8:e69562
Lee SD (2007) Labedella gwakjiensis gen. nov., sp. nov., a novel actinomycete of the family Microbacteriaceae. Int J Syst Evol Microbiol 57:2498–2502
Lee SD, Kim IS (2023) Microbacterium tenebrionis sp. nov. and Microbacterium allomyrinae sp. nov., isolated from larvae of Tenebrio molitor L. and Allomyrina dichotoma, respectively. Int J Syst Evol Microbiol 73:5729
Lee JS, Lee KC, Park YH (2006) Microbacterium koreense sp. nov., from sea water in the South Sea of Korea. Int J Syst Evol Microbiol 56:423–427
Meier-Kolthoff JP, Auch AF, Klenk H-P, Göker M (2013) Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics 14:60
Minnikin DE, Patel PV, Alshamaony L, Goodfellow M (1977) Polar lipid composition in the classification of Nocardia and related bacteria. Int J Syst Bacteriol 27:104–107
Minnikin DE, O’Donnell AG, Goodfellow M, Alderson G, Athalye M et al (1984) An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Methods 2:233–241
Na SI, Kim YO, Yoon SH, Ha SM, Baek I et al (2018) UBCG: up-to-date bacterial core gene set and pipeline for phylogenomic tree reconstruction. J Microbiol 56:280–285
Nouioui I, Carro L, Garcia-Lopez M, Meier-Kolthoff JP, Woyke T et al (2018) Genome-based taxonomic classification of the phylum Actinobacteria. Front Microbiol 9:2007
Orla-Jensen S (1919) The lactic acid bacteria. Høst, Copenhagen
Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW (2014) Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25:1043–1055
Richert K, Brambilla E, Stackebrandt E (2007) The phylogenetic significance of peptidoglycan types: molecular analysis of the genera Microbacterium and Aureobacterium based upon sequence comparison of gyrB, rpoB, recA and ppk and 16S rRNA genes. Syst Appl Microbiol 30:102–108
Richter M, Rósello-Móra R (2009) Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA 106:12196–19131
Saitou N, Nei M (1987) The neighbor-joining method, a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Suzuki K, Hamada M (2012) Genus Microbacterium Orla-Jensen 1919, 179AL emend. Takeuchi and Hatano 1988, 744VP. In: Goodfellow M, Kämpfer P, Hussse H-J, Trujilo M, Suzuki K-I, Ludwig W, William WB (eds) Bergy’s manual of systematic bacteriology, vol 5. Springer, New York, pp 814–855
Takeuchi M, Hatano K (1998) Proposal of six new species in the genus Microbacterium and transfer of Flavobacterium marinotypicum ZoBell and Upham to the genus Microbacterium as Microbacterium maritypicum comb. nov. Int J Syst Bacteriol 48:973–982
Terlouw BR, Blin K, Navarro-Muñoz JC, Avalon NE, Chevrette MG (2023) MIBiG 3.0: a community-driven effort to annotate experimentally validated biosynthetic gene clusters. Nucleic Acids Res 51:D603–D610
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 24:4876–4882
Tian Z, Yang J, Lai XH, Pu J, Jin D et al (2021) Microbacterium caowuchunii sp. nov. and Microbacterium lushaniae sp. nov., isolated from plateau pika (Ochotona curzoniae) on the Qinghai-Tibet Plateau of PR China. Int J Syst Evol Microbiol 71:4662
Uchida K, Aida K (1984) An improved method for the glycolate test for simple identification of the acyl type of bacterial cell walls. J Gen Appl Microbiol 30:131–134
Xie F, Niu S, Lin X, Pei S, Jiang L et al (2021) Description of Microbacterium luteum sp. nov., Microbacterium cremeum sp. nov., and Microbacterium atlanticum sp. nov., three novel C50 carotenoid producing bacteria. J Microbiol 59:886–897
Yoon S-H, Ha S-M, Lim J, Kwon S, Chun J (2017) A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 110:1281–1286
Zhang L, Xi L, Ruan J, Huang Y (2012) Microbacterium marinum sp. nov., isolated from deep-sea water. Syst Appl Microbiol 35:81–85
Acknowledgements
The authors are grateful to Professor Aharon Oren (The Hebrew University of Jerusalem, Israel) for help in preparing species names.
Funding
This work was performed by the financial support of BioPS Co., Ltd.
Author information
Authors and Affiliations
Contributions
SDL performed bacterial isolation and bioinformatic analyses. HLY carried out phenotypic assays and data collection. JSK performed the preparation and analysis of fatty acids. SDL wrote the manuscript. ISK made financial support. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical statement
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Communicated by Yusuf Akhter.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The DDBJ/ENA/GenBank accession numbers for 16S rRNA gene sequences of strains KSW-18T, KSW2-22 and KSW4-11T are OR039656-OR039658, respectively. The DDBJ/ENA/GenBank accession numbers of genome sequences of strains KSW-18T, KSW2-22 and KSW4-11T are JAUZVT010000000-JAUZVV010000000, respectively.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Lee, S.D., Yang, H.L., Kim, JS. et al. Microbacterium aquilitoris sp. nov. and Microbacterium gwkjiense sp. nov., isolated from beach. Arch Microbiol 206, 100 (2024). https://doi.org/10.1007/s00203-023-03804-5
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00203-023-03804-5