Abstract
A genome-based polyphasic study was undertaken to establish the taxonomic status of an actinobacterium strain isolated from an actinorhizal root nodule. Strain ncl1T was found to have chemotaxonomic, cultural and morphological properties characteristic of members of the genus Nocardia. The strain was closely related to Nocardia aurea in the phylogenetic trees based on 16S rRNA gene and genome sequences. The draft genome of the strain is 8.9 Mbp in size, has a genomic DNA G + C content of 67.0% and was predicted to contain at least 19 biosynthetic gene clusters encoding for specialized metabolites. Strain ncl1T was distinguished from its closest neighbour, N. aurea DSM 103986T, by a broad range of phenotypic properties and by low average nucleotide identity and digital DNA-DNA hybridization scores. Consequently, the strain represents a novel Nocardia species for which the name Nocardia noduli sp. nov. is proposed. The type strain is ncl1T (CECT 30123T = DSM 110878T). The present study provides further evidence that actinorhizal nodules are a source of novel species of Nocardia.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Data acquired from polyphasic taxonomic and genome-based studies led to marked improvements in the classification of the genus Nocardia and related mycolic acid-containing bacteria (Nouioui et al. 2018) which had previously been classified using a few subjectively weighted features. The genus forms a monophyletic clade within the family Nocardiaceae (Goodfellow and Maldonaldo 2012) and encompasses 120 validly published species (https://lpsn.dsmz.de/genus/nocardia) most of which have been recognized using combinations of genotypic and phenotypic properties (Nouioui et al. 2020) though the steady flow of new species shows that the genus remains underspeciated (Goodfellow and Maldonaldo 2012; Nouioui et al. 2020).
Nocardia are well known as serious causal agents of progressive invasive infections of humans and animals, notably mycetoma and nocardiosis (Conville et al. 2018), but are also seen to be successful saprophytes as they degrade complex organic compounds (Luo et al. 2014), and as attractive sources of new drug leads (Männle et al. 2020). Nocardia strains have been isolated from diverse ecological settings (Goodfellow and Maldonaldo 2012), notably from soil, sediments, sponges and wastewater treatment plants (Goodfellow and Maldonado 2012), as well as from coastal marine foams (Wright et al. 2021) and healthy plant tissues (Kaewkla and Franco 2010), including nodules of actinorhizal plants (Ghodhbane-Gtari et al. 2014) thereby suggesting they may be beneficial for plant growth (Nouioui et al. 2022).
The improved classification of the genus provides an invaluable framework for the detection of new Nocardia species. The present study was designed to establish the taxonomic status of a Nocardia strain isolated from a sterilized root nodule of Alnus glutinosa and to gain an insight into its biotechnological potential. The isolate ncl1T was the focus of a genomic-based polyphasic study which showed that it represented a new species of the genus Nocardia for which the name Nocardia noduli sp. nov. is proposed. Genome mining showed that the type strain of the species has the potential to synthesize novel natural products, notably antibiotics and anticancer compounds.
Materials and methods
Isolation, maintenance and cultivation
Strain ncl1T was isolated from a surfaced sterilized root nodule of an Alnus glutinosa tree growing in Leazes Park, Newcastle upon Tyne, UK, as described by Nouioui et al. (2022). An active culture of the strain was obtained following growth on yeast extract-malt extract agar (International Streptomyces project [ISP] medium 2 (Shirling and Gottlieb 1966)). The strain was checked for purity and maintained in 35% (w/v) glycerol at − 80 °C, as was the type strain of N. aurea DSM 103986T, which was obtained from the German Collection of Microorganisms and Cell Cultures (DSMZ). Biomass for the chemotaxonomic studies carried out on the strains was harvested from ISP2 broths shaken at 200 rpm in baffled flasks for 7 days at 28 °C. The harvested cells were washed in distilled water and freeze dried.
Cultural and morphological properties of strain ncl1T
Acid-alcohol-fast staining was carried out following growth on ISP2 agar for 10 days at 28 °C using the Ziehl–Neelsen method (Gordon 1967). Hanging drop preparations were examined to check for motility. The micromorphological properties of the strain were observed by light microscopy of the Gram stained smear. The ability of the strain to grow at a range of temperatures (4 °C, 10 °C, 15 °C, 25 °C, 28 °C, 37 °C, 42 °C and 45 °C) and pH (pH4.0–9.0 at intervals of 0.50 unit) was recorded from ISP2 agar plates. The pH levels were achieved using KH2PO4/HCl, KH2PO4/K2HPO4 and K2HPO4/NaOH buffer systems. Growth and cultural properties were determined after incubation for 7 days at 28 °C on GYM agar (DSMZ medium 65), nutrient agar (NA, (MacFaddin 1986)), peptone-meat extract-glucose agar (DSMZ medium 250), tryptic soy agar (TSA, (MacFaddin 1986)) and ISP media 2, 3, 4 (Shirling and Gottlieb 1966). The ability of the strain to grow under anaerobic conditions was determined by inoculating it onto ISP2 agar and incubating at 28 °C for 7 days using anaerobic bag system (Sigma-Aldrich 68,061).
Molecular identification and genome sequencing
DNA was extracted from strain ncl1T prior to PCR-mediated amplification of a 16S rRNA gene (Weisburg et al. 1991) and sequencing using the Sanger method (Sanger and Coulson 1975; Sanger et al. 1977). Genomic DNA of the strain was extracted, purified and quantified following the protocol provided by MicrobesNG (Birmingham, UK, https://microbesng.com/microbesng-faq/), then sequenced on an Illumina HiSeq 250 next generation sequencing platform using the 250 bp paired end sequencing protocol at MicrobesNG. Assembly of the reads and associated procedures were also carried out at MicrobesNG. The draft genome sequence of the strain was annotated using the RAST-SEED webserver (Aziz et al. 2008). The completeness of the genome sequence of the strain was estimated using BUSCO v.5.1.2 (Seppey et al. 2019).
Genome comparison
The draft genome of strain ncl1T was compared with that of N. aurea SYSU K10002T (accession number NZ_QCYJ00000000.1) for several genomic features, notably genome size and digital DNA G + C content. Average nucleotide identity (ortho ANI; Lee et al. 2016), average amino acid identity (AAI) (Konstantinidis and Tiedje 2005) and digital DNA-DNA hybridization (dDDH) (Meier-Kolthoff et al. 2013) similarities were determined from the draft genomes using the ANI calculators from the EzBioCloud (https://www.ezbiocloud.net/tools/ani) and the Genome-to-Genome Distance Calculator GGDC 2.1 (http://ggdc.dsmz.de) webserver, respectively.
Phylogeny
An almost full length 16S rRNA gene sequence extracted directly from the draft genome of strain ncl1T (1523 pb) was deposited in the GenBank under accession number OK597210. This sequence was identical to the 16S rRNA gene sequence obtained using the Sanger method. The gene sequence was compared with corresponding gene sequences of the type strains of closely related Nocardia species retrieved from the EzBioCloud database (Yoon et al. 2017). 16S rRNA gene and whole-genome phylogenetic trees were inferred using the Type Strain Genome Server (TYG), available on the GGDC webpage (Meier-Kolthoff and Göker 2019).
Phenotypic and chemotaxonomic properties
Strain ncl1T and N. aurea DSMZ 103986T were examined for a range of phenotypic features; notably for their ability to use sole carbon and nitrogen sources; to grow at various pH values and in the presence of antibiotics and inorganic inhibitory compounds using GENIII microplates and an Omnilog device (Biolog Inc., Hayward, CA, USA), as described by Nouioui et al. (2020). The resultant data were analysed using opm package version 1.3.36 (Vaas et al. 2013). Enzymatic profiles of the strains were established using API-ZYM kits (BioMerieux, Lyon, France) by following the manufacturer’s instructions. Catalase and oxidase tests were determined using standard procedures. All of the tests cited above were performed in duplicate.
Standard chromatographic procedures were used to establish the polar lipid profile and diaminopimelic acid isomers using the integrated method of Minnikin et al. (1984) and the protocol of Schleifer and Kandler (1972), respectively. Whole-organism sugar patterns were determined after Lechevalier and Lechevalier (1970) and Staneck and Roberts (1974). Mycolic acids were extracted and purified as described by Goodfellow et al. (1976) and identified by gas chromatography (Agilent Technologies 6890 N, Santa Clara, CA, USA).
Cellular fatty acids extracted from the strain and N. aurea DSM 103986T were methylated following Miller (1982), as modified by Kuykendall et al. (1988) and analysed by gas chromatography using the Agilent instrument. The resultant peaks were identified using the standard Microbial Identification (MIDI) system version 4.5 and the ACTIN6 database (Sasser 1990).
Biosynthetic gene clusters coding for specialized metabolites
The presence of natural product-biosynthetic gene clusters (NP-BGCs) in the genome of the isolate and the N. aurea strain were detected using antiSMASH, version 6.0 (Blin et al. 2019) with the detection relaxed option.
Results and discussion
Cultural, chemotaxonomic and biochemical properties
Strain ncl1T did not grow under anaerobic conditions, but did grow from 28 °C to 37 °C, optimally at 28 °C, and from pH 6–7.5. It formed orange colonies on GYM and ISP2 media and orange yellowish aerial hyphae on DSM 250 medium. It was also found to be catalase positive and oxidase negative.
Excellent congruence was found between the phenotypic data acquired on the duplicated tests. Table 1 shows that the isolate and N. aurea DSM 103986T can be distinguished using a combination of phenotypic features though they also have many properties in common. Only the isolate was able to metabolise d-arabitol, D-fucose, β-gentiobiose, glucuronamide, N-acetyl-D-glucosamine, d-mannitol, methyl pyruvate and l-rhamnose (sugars), gelatin (polymer), and was resistant to vancomycin. In contrast, N. aurea, unlike the strain, metabolised 3-O-methyl-d-glucose, d-turanose, β-methyl-d-glucoside, and d-salicin (sugars) and was resistant to fusidic acid and minocycline.
Chemotaxonomic, colonial and micromorphological features of the strain were consistent with its classification in the genus Nocardia (Goodfellow and Maldonaldo 2012). The strain was found to be aerobic, nonmotile, Gram-stain-positive, partial acid-alcohol-fast and formed substrate and aerial hyphae that fragmented into rod and coccoid-like elements. It was shown to have meso-A2pm as the wall peptidoglycan, arabinose, galactose and glucose as whole-organism sugars; mycolic acids with 46–64 carbon atoms and a polar lipid profile composed of diphosphatidylglycerol, phosphatidylethanolamine (diagnostic phospholipid) and phosphatidylinositol and spots corresponding to unknown lipid, phosphoglycolipid and glycolipid components (Figure S1). The strain and the N. aurea strain were found to be rich in straight chain, saturated, unsaturated and 10-methyl (tuberculostearic) fatty acids though quantitative differences were found between the various components (Table S1). Both strains feature meso-A2pm as the wall peptidoglycan and arabinose, galactose and glucose as diagnostic sugars, but can be distinguished as the strain also contain glucose and the N. aurea strain ribose (Fang et al. 2019).
Genome comparison
The draft genome sequence of the strain has been deposited in the GenBank database under accession number JAFMPL000000000. The genome of the strain has a BUSCO score of 99.7% complete genes (355/356 from the actinobacteria_class_odb10). The genome sizes of strain ncl1T and the type strain of N. aurea were 8.9 Mbp and 8.3 Mbp, respectively, and the corresponding genomic DNA G + C contents were 67.0 and 67.4%. The dDDH similarity between the strains was 61.7% (Table 2), a value much lower than the corresponding threshold of 70% used to assign closely related strains to the same species (Wayne et al. 1987). However, the ANI value between these strains was shown to be 95%, and the AAI 93.7%, which is below the cut-off point of 95–96% for species demarcation (Goris et al. 2007; Richter and Rosselló-Móra 2009; Thompson et al. 2013).
Phylogeny
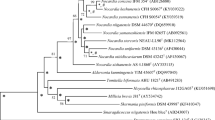
The maximum-likelihood 16S rRNA Nocardia tree (Fig. 1) showed that the strain formed a well separated branch in a clade that included the type strains of seven Nocardia species. Its closest relative was found to be N. aurea SYSU K10002T (99.4%) in the phylogenomic tree. Greater confidence can be placed in the topology of phylogenomic trees as they are generated from millions of unit characters not limited to about 1500 as in the case of corresponding 16S rRNA gene trees (Nouioui et al. 2018). It is evident from the phylogenomic tree that the strain and N. aurea SYSU K10002T formed a well-supported subclade that is most closely related to a second well delineated taxon composed of Nocardia arizonensis NBRC 108935T and Nocardia takedensis NBRC 100417T (Fig. 2). The resultant well-supported clade was seen to be distant from corresponding clades encompassing several Nocardia species, notably Nocardia brasiliensis NBRC 14402T and Nocardia vulneris NBRC 108936T, and N. altamirensis NBRC 108246T and Nocardia tenerifensis DSM 44704T, respectively.
Biosynthetic gene clusters coding for specialized metabolites
AntiSMASH predicts NP-BGCs and potential products based on the percentage of genes from the closest known bioclusters which show BLAST hits in the genomes of strains of interest. In the present study, the genome of strain ncl1T and the N. aurea DSM 103986T contained 19 and 31 BGCs, respectively. Many of the bioclusters were predicted to encode for different classes of specialized metabolites, notably non-ribosomal peptide synthases (NRPS); type 1 and 2 polyketide synthases (PKS) and terpenes (Table 3), thereby providing further evidence that nocardiae have the capacity to produce diverse natural products (Männle et al. 2020). The genome of each strain contained bioclusters associated with the synthesis of nocobactin (87% gene identity), a UV-active siderophore first reported from cultures of Nocardia asteroides (Ratledge and Snow 1974), and streptobactin (11% gene identity), a tricatechol-type siderophore from a marine-derived Streptomyces strain which showed iron-chelating activity (Matsuo et al. 2011). The strains were also found to have bioclusters predicted to encode for the volatile terpenes geosmin (100% gene identity), isorenieratene (57% gene identity), 2-methylisoborneol (75% gene similarity), and acyldepsipeptide (10% gene identity). The latter was isolated from a marine-derived Streptomyces atratus strain and was found to be active against Mycobacterium tuberculosis (Sun et al. 2019). The strains were found to have the capacity to produce caniferolides A to D with 12% to 14% gene identity, respectively. The latter are PKS compounds which were initially detected in Streptomyces caniferus and shown to inhibit the growth of Aspergillus spp. and Candida albicans (Pérez-Victoria et al. 2019); caniferolide A has been used to treat Alzheimer’s disease (Alvariño et al. 2019).
Many of the remaining bioclusters found in the genomes of the two strains were found to be strain specific (Table 3). Strain ncl1T, for instance, contained BGCs predicted to encode for atratumycin (5% gene identity), cinerubin B (14% gene identity), an anthracycline antibiotic produced by a Streptomyces strain which has antitumor properties (Silva et al. 2020); cyphomycin (2% gene identity), a PKS from a marine-derived Streptomyces strain which is used to control multi-drug resistant fungal pathogens (Chevrette et al. 2019); polyoxypeptin A, which exhibits potent apoptosis-inducing activity towards human pancreatic carcinomic cells (Du et al. 2014), and simocyclinone D8, a cytostatic angucyclinone antibiotic from a Streptomyces strain which is active against Gram-stain-positive bacteria and tumour cell lines (Schimana et al. 2000).
In contrast, only the N. aurea strain has bioclusters associated with the synthesis of cosmomycin D (5% gene identity), an anthracycline antibiotic which shows antitumor activity (Ando et al. 1985); platencin (6% gene identity), a betalactone which inhibits multi-drug resistant M. tuberculosis strains (Martens and Demain 2011); pepticinnamin E (6% gene identity), a farnesyl transferase inhibitor used in cancer, malaria, and trypanosome therapy (Santa et al. 2019); xantholipin, a polycyclic xanthone antibiotic derived from Streptomyces flavogriseus, which was found to be cytotoxic and to show antibacterial activity against Gram-positive bacteria, including methicillin-resistant Staphylococcus aureus and vancomycin-resistant Enterococcus faecalis (Wu et al. 2017). In addition, the aziridine alkaloid antibiotic ficellomycin (3% gene identity) and the type I PKs compounds tetarimycin A and B (3% gene identity) inhibit multi-drug resistant S. aureus strains (Kallifidas et al. 2012; He et al. 2018). The N. aurea strain also has the capacity to produce abyssomicin C/ atrop-abyssomicin C (25% gene similarity), polycyclic macrolactones, first detected in the type strain of Verrucosispora maris (now Micromonospora maris) (Nouioui et al. 2018), which show pronounced activity against multi-drug resistant Gram-positive bacteria (Fiedler 2021). Additional compounds associated with each of the strains are shown in Table 3.
It is evident from the results outlined above that strain ncl1T and N. aurea DSM 103986T, its closest phylogenomic neighbour, have genomes rich in BGCs predicted to encode for novel polyketides. These results provide further evidence that nocardiae should feature more prominently in natural product discovery campaigns designed to find novel antibiotics of therapeutic value. This prospectus is in line with the view that novel Nocardia species represent a prolific source of natural products, one that rivals that of better characterised genera such as Amycolatopsis and Streptomyces (Männle et al. 2020).
Conclusions
Nodular tissues provide shelter and a plentiful supply of carbohydrates for bacteria, including actinobacteria found to belong to the genera Frankia, Micromonospora and Streptomyces. The present study provides further evidence that novel Nocardia species are associated with actinorhizal nodules though ecophysiological studies are needed to establish their ecological role and potential as inoculants to promote plant growth. Further, their ability to inhibit phytopathogens offers a role as an eco-friendly alternative to the use of pesticides in agriculture.
It can be concluded from the phylogenetic and phylogenomic trees that the strain forms a distinct lineage that is most closely related to the type strain of N. aurea. However, it can be distinguished from the latter using a broad range of phenotypic data and by low ANI and dDDH similarities. It is, therefore, proposed that strain ncl1T represents a new species within the genus Nocardia for which the name Nocardia noduli sp. nov. is proposed.
Description of Nocardia noduli. sp. nov
Nocardia noduli (no’du.li. L. gen. dim. n. noduli, of a nodule, referring to the isolation of the type strain from a root nodule of Alnus glutinosa).
Aerobic, Gram-stain-positive, non-motile organism which forms substrate and aerial hyphae that fragment into rod-and coccoid-like elements. Orange colonies are formed on GYM and ISP2 agar. Grows from pH 5–7.5, optimally at pH 7.0 and from 28 to 37 °C, optimally at 28 °C, and in presence of up to 4% w/v sodium chloride. It is catalase positive, oxidase negative, able to metabolise D-arabitol, D-fucose, β-gentiobiose, glucuronamide, N-acetyl-d-glucosamine, D-mannitol, methyl pyruvate and L-rhamnose (sugars), gelatin (polymer), is resistant to vancomycin and positive for α-galactosidase, valine aminopeptidase, β-glucosidase, n-acetyl-β-glucosaminidase and naphthol-AS-BI phosphohydrolase. The diamino acid of the peptidoglycan is meso-A2pm, the whole-organism sugars are arabinose, galactose and glucose; the predominant fatty acids (> 10%) are C16:0, C18:1 ω9c and 10-methyl C18:0, and the mycolic acids have 46–64 carbon atoms. The polar lipids consist of diphosphatidylglycerol, phosphatidylethanolamine, phosphatidylinositol, a glycophospholipid and two unidentified components. The genome size of the strain is 8.9 Mbp and its genomic DNA G + C content 67.0%. The genome is rich in biosynthetic gene clusters predicted to encode for specialized metabolites, notably antibiotics.
The type strain, ncl1T (CECT 30123T = DSM 110878T), was isolated from the root nodule of an Alnus glutinosa plant growing in Leazes Park, Newcastle upon Tyne, UK.
The 16S rRNA gene and genome sequences have been deposited in the DDBJ/ENA/GenBank databases under the accession numbers OK597210 and JAFMPL000000000, respectively.
The Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accession number JAFMPL000000000. The version described in this paper is version JAFMPL010000000.
References
Alvariño R, Alonso E, Lacret R, Oves-Costales D, Genilloud O et al (2019) Caniferolide A, a macrolide from Streptomyces caniferus, attenuates neuroinflammation, oxidative stress, amyloid-beta, and tau pathology in vitro. Mol Pharm 16:1456–1466. https://doi.org/10.1021/acs.molpharmaceut.8b01090
Ando T, Hirayama K, Takahashi R, Horino I, Etoh Y et al (1985) Cosmomycin D, a new anthracycline antibiotic. Agric Biol Chem 49:259–262. https://doi.org/10.1271/bbb1961.49.259
Aziz RK, Bartels D, Best AA, DeJongh M, Disz T et al (2008) The RAST Server: rapid annotations using subsystems technology. BMC Genom 9:75. https://doi.org/10.1186/1471-2164-9-75
Blin K, Shaw S, Steinke K, Villebro R, Ziemert N et al (2019) antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res 47:W81–W87. https://doi.org/10.1093/nar/gkz310
Chevrette MG, Carlson CM, Ortega HE, Thomas C, Ananiev GE et al (2019) The antimicrobial potential of Streptomyces from insect microbiomes. Nat Commun 10:516. https://doi.org/10.1038/s41467-019-08438-0
Conville PS, Brown-Elliott BA, Smith T, Zelazny AM (2018) The complexities of Nocardia taxonomy and identification. J Clin Microbiol 56:e01419-e1517. https://doi.org/10.1128/JCM.01419-17
Du Y, Wang Y, Huang T, Tao M, Deng Z et al (2014) Identification and characterization of the biosynthetic gene cluster of polyoxypeptin A, a potent apoptosis inducer. BMC Microbiol 14:30. https://doi.org/10.1186/1471-2180-14-30
Fang B-Z, Han M-X, Zhang L-Y, Jiao J-Y, Zhang X-T et al (2019) Nocardia aurea sp. nov., a novel actinobacterium isolated from a karstic subterranean environment. Int J Syst Evol Microbiol 69:159–164. https://doi.org/10.1099/ijsem.0.003122
Fiedler H-P (2021) Abyssomicins: a 20-year retrospective view. Mar Drugs 19:299. https://doi.org/10.3390/md19060299
Ghodhbane-Gtari F, Nouioui I, Salem K, Ktari A, del Montero-Calasanz M et al (2014) Nocardia casuarinae sp. nov., an actinobacterial endophyte isolated from root nodules of Casuarina glauca. Antonie Van Leeuwenhoek 105:1099–1106. https://doi.org/10.1007/s10482-014-0168-6
Goodfellow M, Maldonaldo LA (2012) Genus I. Nocardia Trevisan 1889al. In: Goodfellow M, Kämpfer P, Busse H-J, Trujillo ME, Suzuki K-I, Ludwig W, Whitman WB (eds) Bergey’s manual of systematic bacteriology. 2nd ed, vol 5. The Actinobacteria Parts A and B. Springer, New York, pp 376–418
Goodfellow M, Collins MD, Minnikin DE (1976) Thin-layer chromatographic analysis of mycolic acid and other long-chain components in whole-organism methanolysates of coryneform and related taxa. J Gen Microbiol 96:351–358. https://doi.org/10.1099/00221287-96-2-351
Gordon RE (1967) The taxonomy of soil bacteria. In: Grey TRG, Parkinson D (eds) The ecology of soil bacteria. Liverpool University Press, Liverpool
Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P et al (2007) DNA–DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol 57:81–91. https://doi.org/10.1099/ijs.0.64483-0
He X, Li M, Song S, Wu X, Zhang J et al (2018) Ficellomycin: an aziridine alkaloid antibiotic with potential therapeutic capacity. Appl Microbiol Biotechnol 102:4345–4354. https://doi.org/10.1007/s00253-018-8934-4
Kaewkla O, Franco CMM (2010) Nocardia callitridis sp. nov., an endophytic actinobacterium isolated from a surface-sterilized root of an Australian native pine tree. Int J Syst Evol Microbiol 60:1532–1536. https://doi.org/10.1099/ijs.0.016337-0
Kallifidas D, Kang H-S, Brady SF (2012) Tetarimycin A, an MRSA active antibiotic identified through induced expression of environmental DNA gene clusters. J Am Chem Soc 134:19552–19555. https://doi.org/10.1021/ja3093828
Konstantinidis KT, Tiedje JM (2005) Genomic insights that advance the species definition for prokaryotes. Proc Natl Acad Sci USA 102:2567–2572. https://doi.org/10.1073/pnas.0409727102
Kuykendall LD, Roy MA, O'Neill JJ, Devine TE (1988) Fatty acids, antibiotic resistance, and deoxyribonucleic acid homology groups of Bradyrhizobiurn japonicum. Int J Syst Evol Microbiol 38:358-361. https://doi.org/10.1099/00207713-38-4-358
Lechevalier MP, Lechevalier H (1970) Chemical composition as a criterion in the classification of aerobic actinomycetes. Int J Syst Evol Microbiol 20:435–443. https://doi.org/10.1099/00207713-20-4-435
Lee I, Ouk Kim Y, Park S-C, Chun J (2016) OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol 66:1100–1103. https://doi.org/10.1099/ijsem.0.000760
Luo Q, Hiessl S, Poehlein A, Daniel R, Steinbüchel A (2014) Insights into the microbial degradation of rubber and gutta-percha by analysis of the complete genome of Nocardia nova SH22a. Appl Environ Microbiol 80:3895–3907. https://doi.org/10.1128/AEM.00473-14
MacFaddin JF (1986) Media for isolation cultivation identification - maintenance of medical bacteria. Baltimore, London. https://doi.org/10.1002/jobm.3620260414
Männle D, McKinnie SMK, Mantri SS, Steinke K, Lu Z et al (2020) Comparative genomics and metabolomics in the genus Nocardia. mSystems 5:e00125-20. https://doi.org/10.1128/mSystems.00125-20
Martens E, Demain AL (2011) Platensimycin and platencin: promising antibiotics for future application in human medicine. J Antibiot (tokyo) 64:705–710. https://doi.org/10.1038/ja.2011.80
Matsuo Y, Kanoh K, Jang J-H, Adachi K, Matsuda S et al (2011) Streptobactin, a tricatechol-type siderophore from marine-derived Streptomyces sp. YM5-799. J Nat Prod 74:2371–2376. https://doi.org/10.1021/np200290j
Meier-Kolthoff JP, Göker M (2019) TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat Commun 10:2182. https://doi.org/10.1038/s41467-019-10210-3
Meier-Kolthoff JP, Auch AF, Klenk H-P, Göker M (2013) Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform 14:60. https://doi.org/10.1186/1471-2105-14-60
Miller LT (1982) Single derivatization method for routine analysis of bacterial whole-cell fatty acid methyl esters, including hydroxy acids. J Clin Microbiol 16:584–586. https://doi.org/10.1128/jcm.16.3.584-586.1982
Minnikin DE, O’Donnell AG, Goodfellow M, Alderson G, Athalye M et al (1984) An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Methods 5:233–241. https://doi.org/10.1016/0167-7012(84)90018-6
Nouioui I, Carro L, García-López M, Meier-Kolthoff JP, Woyke T et al (2018) Genome-based taxonomic classification of the phylum Actinobacteria. Front Microbiol 9:2007. https://doi.org/10.3389/fmicb.2018.02007
Nouioui I, Cortés-Albayay C, Neumann-Schaal M, Vicente D, Cilla G et al (2020) Genomic virulence features of two novel species Nocardia barduliensis sp. nov. and Nocardia gipuzkoensis sp. nov., isolated from patients with chronic pulmonary diseases. Microorganisms 8:1517. https://doi.org/10.3390/microorganisms8101517
Nouioui I, Ha S-m, Baek I, Chun J, Goodfellow M (2022) Genome insights into the pharmaceutical and the plant growth promoting features of the novel species, Nocardia alni sp. nov. BMC Genom 23:70. https://doi.org/10.1186/s12864-021-08257-y
Pérez-Victoria I, Oves-Costales D, Lacret R, Martín J, Sánchez-Hidalgo M et al (2019) Structure elucidation and biosynthetic gene cluster analysis of caniferolides A-D, new bioactive 36-membered macrolides from the marine-derived Streptomyces caniferus CA-271066. Org Biomol Chem 17:2954–2971. https://doi.org/10.1039/c8ob03115k
Ratledge C, Snow GA (1974) Isolation and structure of nocobactin NA, a lipid-soluble iron-binding compound from Nocardia asteroides. Biochem J 139:407–413. https://doi.org/10.1042/bj1390407
Richter M, Rosselló-Móra R (2009) Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA 106:19126–19131. https://doi.org/10.1073/pnas.0906412106
Sanger F, Coulson AR (1975) A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J Mol Biol 94:441–448. https://doi.org/10.1016/0022-2836(75)90213-2
Sanger F, Nicklen S, Coulson AR (1977) DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA 74:5463–5467. https://doi.org/10.1073/pnas.74.12.5463
Santa Maria KC, Chan AN, O’Neill EM, Li B (2019) Targeted rediscovery and biosynthesis of the farnesyl-transferase inhibitor pepticinnamin E. ChemBioChem Commun 20:1387–1393. https://doi.org/10.1002/cbic.201900025
Sasser M (1990) Bacterial identification by gas chromatographic analysis of fatty acid methyl esters (GC-FAME). Technical note 101. Microbial ID USA Inc, Newark, p 6
Schimana J, Fiedler H-P, Groth I, Süssmuth R, Beil W et al (2000) Simocyclinones, novel cytostatic angucyclinone antibiotics produced by Streptomyces antibioticus Tü 6040I. Taxonomy, fermentation, isolation and biological activities. J Antibiot (tokyo) 53:779–787. https://doi.org/10.7164/antibiotics.53.779
Schleifer KH, Kandler O (1972) Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol Rev 36:407–477. https://doi.org/10.1128/br.36.4.407-477.1972
Seppey M, Manni M, Zdobnov EM (2019) BUSCO: assessing genome assembly and annotation completeness. In: Kollmar M (ed) Gene prediction: methods and protocols. Springer, New York, pp 227–245. https://doi.org/10.1007/978-1-4939-9173-0_14
Shirling EB, Gottlieb D (1966) Methods for characterization of Streptomyces species. Int J Syst Evol Microbiol 16:313–340. https://doi.org/10.1099/00207713-16-3-313
Silva LJ, Crevelin EJ, Souza DT, Lacerda-Júnior GV, de Oliveira VM et al (2020) Actinobacteria from Antarctica as a source for anticancer discovery. Sci Rep 10:13870. https://doi.org/10.1038/s41598-020-69786-2
Staneck JL, Roberts GD (1974) Simplified approach to identification of aerobic actinomycetes by thin-layer chromatography. Appl Microbiol 28:226–231. https://doi.org/10.1128/am.28.2.226-231.1974
Sun C, Yang Z, Zhang C, Liu Z, He J et al (2019) Genome mining of Streptomyces atratus SCSIO ZH16: discovery of Atratumycin and identification of its biosynthetic gene cluster. Org Lett 21:1453–1457. https://doi.org/10.1021/acs.orglett.9b00208
Thompson CC, Chimetto L, Edwards RA, Swings J, Stackebrandt E, Thompson FL (2013) Microbial genomic taxonomy. BMC Genomics 14:913. https://doi.org/10.1186/1471-2164-14-913
Vaas LAI, Sikorski J, Hofner B, Fiebig A, Buddruhs N et al (2013) opm: an R package for analysing OmniLog® phenotype microarray data. Bioinformatics 29:1823–1824. https://doi.org/10.1093/bioinformatics/btt291
Wayne LG, Brenner DJ, Colwell RR, Grimont PAD, Kandler O et al (1987) Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Evol Microbiol 37:463–464. https://doi.org/10.1099/00207713-37-4-463
Weisburg WG, Barns SM, Pelletier DA, Lane DJ (1991) 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173:697–703. https://doi.org/10.1128/jb.173.2.697-703.1991
Wright L, Katouli M, Kurtböke Dİ (2021) Isolation and characterization of nocardiae associated with foaming coastal marine waters. Pathogens 10:579. https://doi.org/10.3390/pathogens10050579
Wu S, Huang T, Xie D, Wo J, Wang X et al (2017) Xantholipin B produced by the stnR inactivation mutant Streptomyces flocculus CGMCC 4.1223 WJN-1. J Antibiot 70:90–95. https://doi.org/10.1038/ja.2016.60
Yoon S-H, Ha S-M, Kwon S, Lim J, Kim Y et al (2017) Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol 67:1613–1617. https://doi.org/10.1099/ijsem.0.001755
Acknowledgements
We are grateful to Dr. Katharina Huber, Dr. Meina Neumann-Schaal and Ms. Caroline Pilke (Leibniz Institute DSMZ–German Collection of Microorganisms and Cell Cultures, 38124 Braunschweig, Germany), for their support with some phenotypic analyses and to Dr. Juan P. Gomez Escribano (DSMZ), for support in estimating completeness of the genome. Genome sequencing was provided by MicrobesNG (http://www.microbesng.com) which is supported by the BBSRC (grant number BB/L024209/1).
Funding
Open Access funding enabled and organized by Projekt DEAL. The authors declare that no funds, grants, or other support were received during the preparation of this manuscript.
Author information
Authors and Affiliations
Contributions
IN and MG conceived the study. Material preparation, data collection and analysis were performed by all authors. The first draft of the manuscript was written by IN and MG and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Additional information
Communicated by Erko Stackebrandt.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Nouioui, I., Pötter, G., Jando, M. et al. Nocardia noduli sp. nov., a novel actinobacterium with biotechnological potential. Arch Microbiol 204, 260 (2022). https://doi.org/10.1007/s00203-022-02878-x
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00203-022-02878-x