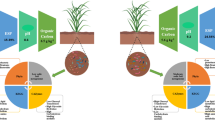
Abstract
Kappaphycus alvarezii seaweed extract (KSWE) is known to enhance crop productivity and impart stress tolerance. Close to one quarter of foliar spray applied to maize falls on the soil, either as drift or from leaf as drip. It was hypothesized that the drift spray would profoundly influence soil microbes under stress. An experiment was conducted with five treatments, with or without KSWE application at critical stages of maize grown under soil moisture stress and compared with an irrigated control. An Illumina platform was employed for the analysis of the V3–V4 region of 16S rRNA gene from the soil metagenome. A total of 345,552 operational taxonomic units were generated which were classified into 55 phyla, 152 classes, 240 orders, 305 families and 593 genera. Shannon’s index and Shannon’s equitability indicated increased soil bacterial diversity after multiple KSWE applications under conditions of abiotic duress. The abundance of the genera Alicyclobacillus, Anaerolinea, Bacillus, Balneimonas, Nitrospira, Rubrobacter and Steroidobacter decreased (49–79%) under drought imposed at the V5,10 and 15 stages of maize over the irrigated control, while it significantly improved when followed by KSWE application under drought. Flavobacterium, Nitrosomonas, Nitrosovibrio, Rubrobacter genera and several other bacterial taxa which are important for plant growth promotion and nutrient cycling were found to be enriched by KSWE application under drought conditions. Treatments having enriched microbial abundance due to KSWE application under stress recorded higher soil enzymatic activities and plant cob yield, suggesting the contribution of altered soil ecology mediated by KSWE as one of the reasons for improvement of yield.







Similar content being viewed by others
Availability of data and material
The datasets generated during and/or analysed during the current study are available in the National Center for Biotechnology Information (NCBI) repository, https://trace.ncbi.nlm.nih.gov/Traces/sra/?study=SRP180107
Code availability
Not applicable.
References
Achal V, Pan X, Fu Q, Zhang D (2012) Biomineralization based remediation of As (III) contaminated soil by Sporosarcina ginsengisoli. J Hazard Mater 201:178–184. https://doi.org/10.1016/j.jhazmat.2011.11.067
Alam MZ, Braun G, Norrie J, Hodges DM (2013) Effect of Ascophyllum extract application on plant growth, fruit yield and soil microbial communities of strawberry. Can J Plant Sci 93:23–36. https://doi.org/10.4141/cjps2011-260
Amiraux R, Rontani JF, Armougom F, Frouin E, Babin M, Artigue L, Bonin P (2021) Bacterial diversity and lipid biomarkers in sea ice and sinking particulate organic material during the melt season in the Canadian Arctic. Elem Sci Anthr 9:1. https://doi.org/10.1525/elementa.2021.040
Anand KGV, Kubavat D, Trivedi K, Agarwal PK, Wheeler C, Ghosh A (2015) Long-term application of Jatropha press cake promotes seed yield by enhanced soil organic carbon accumulation, microbial biomass and enzymatic activities in soils of semi-arid tropical wastelands. Eur J Soil Biol 69:57–65. https://doi.org/10.1016/j.ejsobi.2015.05.005
Aravindraja C, Viszwapriya D, Pandian SK (2013) Ultradeep 16S rRNA sequencing analysis of geographically similar but diverse unexplored marine samples reveal varied bacterial community composition. PLoS ONE 8:e76724. https://doi.org/10.1371/journal.pone.0076724
Berry A, Janssens D, Hümbelin M, Jore JPM, Hoste B, Cleenwerck I, Vancanneyt M et al (2003) Paracoccus zeaxanthinifaciens sp. nov., a zeaxanthin-producing bacterium. Int J Syst Evol Microbiol 53:231–238. https://doi.org/10.1099/ijs.0.02368-0
Boor KJ (2006) Bacterial stress responses: what doesn’t kill them can make them stronger. PLoS Biol 4:e23. https://doi.org/10.1371/journal.pbio.0040023
Boswell CD, Dick RE, Eccles H, Macaskie LE (2001) Phosphate uptake and release by Acinetobacter johnsonii in continuous culture and coupling of phosphate release to heavy metal accumulation. J Ind Microbiol Biotechnol 26:333–340. https://doi.org/10.1038/sj.jim.7000139
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N et al (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335. https://doi.org/10.1038/nmeth.f.303
Castillo AM, Gutiérrez MC, Kamekura M, Xue Y, Ma Y, Cowan DA, Jones BE, Grant WD, Ventosa A (2007) Halovivax ruber sp. nov., an extremely halophilic archaeon isolated from Lake Xilinhot, Inner Mongolia China. Int J Syst Evol Microbiol 57:1024–1027. https://doi.org/10.1099/ijs.0.64899-0
Chuah XQ, Mun W, Teo SS (2017) Comparison study of anti-microbial activity between crude extract of Kappaphycus alvarezii and Andrographis paniculata. Asian Pac J Trop Biomed 7:729–731. https://doi.org/10.1016/j.apjtb.2017.07.003
D’Argenio V, Casaburi G, Precone V, Salvatore F (2014) Comparative metagenomic analysis of human gut microbiome composition using two different bioinformatic pipelines. Biomed Res Int. https://doi.org/10.1155/2014/325340
Dalal A, Bourstein R, Haish N, Shenhar I, Wallach R, Carpentier SC (2019) Dynamic physiological phenotyping of drought-Stressed pepper plants treated with “productivity-enhancing” and “survivability-enhancing” biostimulants. Front Plant Sci 10:905. https://doi.org/10.3389/fpls.2019.00905
Davet P (2004) Microbial ecology of the soil and plant growth. Enfield Science Publishers, New Hampshire
de Podestá GS, de Freitas LG, Dallemole-Giaretta R, Zooca RJF, de Caixeta L, B, Ferraz S, (2013) Meloidogyne javanica control by Pochonia chlamydosporia, Gracilibacillus dipsosauri and soil conditioner in tomato. Summa Phytopathol 39:122–125. https://doi.org/10.1590/S0100-54052013000200007
DeSantis TZ, Hugenholtz P, Keller K, Brodie EL, Larsen N, Piceno YM, Phan R et al (2006a) NAST: a multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res 34:394–399. https://doi.org/10.1093/nar/gkl244
DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T et al (2006b) Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72:5069–5072. https://doi.org/10.1128/AEM.03006-05
Dortet L, Legrand P, Soussy CJ, Cattoir V (2006) Bacterial identification, clinical significance, and antimicrobial susceptibilities of Acinetobacter ursingii and Acinetobacter schindleri, two frequently misidentified opportunistic pathogens. J Clin Microbiol 44:4471–4478. https://doi.org/10.1128/JCM.01535-06
El-Tarabily KA (2006) Rhizosphere-competent isolates of streptomycete and non-streptomycete actinomycetes capable of producing cell-wall-degrading enzymes to control Pythium aphanidermatum damping-off disease of cucumber. Botany 84:211–222. https://doi.org/10.1139/b05-153
Eswaran K, Ghosh PK, Siddhanta AK, Patolia JS, Periyasamy C, Mehta AS, Mody KH, Ramavat BK, Prasad K, Rajyaguru MR, Kulandaivel S, Reddy CRK, Pandya JB, Tewari A (2005) Integrated method for production of carrageenan and liquid fertilizer from fresh seaweeds. United States patent US 6893479
Figueiredo MVB, Martinez CR, Burity HA, Chanway CP (2008) Plant growth-promoting rhizobacteria for improving nodulation and nitrogen fixation in the common bean (Phaseolus vulgaris L.). World J Microbiol Biotechnol 24:1187–1193. https://doi.org/10.1007/s11274-007-9591-4
Galand PE, Casamayor EO, Kirchman DL, Lovejoy C (2009) Ecology of the rare microbial biosphere of the Arctic Ocean Proc Natl Acad. Sci 106:22427–22432. https://doi.org/10.1073/pnas.0908284106
Garai S, Brahmachari K, Sarkar S, Kundu R, Pal M, Pramanick B (2019) Crop growth and productivity of rainy maize-garden pea copping sequence as influenced by Kappaphycus and Gracilaria saps at alluvial soil of West Bengal India. Curr J Appl Sci Technol. https://doi.org/10.9734/CJAST/2019/v36i230227
Garrity GM (2005) Systematic bacteriology. Proteobacteria, Part C Alpha-, Beta-, Delta-, Epsilonproteobacteria, Bergey’s Manual Trust, Department of Microbiology and Molecular Genetics 2
Geng C, Nie X, Tang Z, Zhang Y, Lin J, Sun M, Peng D (2016) A novel serine protease, Sep1, from Bacillus firmus DS-1 has nematicidal activity and degrades multiple intestinal-associated nematode proteins. Sci Rep 6:1–12. https://doi.org/10.1038/srep25012
Ghosh A, Vijay Anand KG, Seth A (2015) Life cycle impact assessment of seaweed based biostimulant production from onshore cultivated Kappaphycus alvarezii (Doty) Doty ex Silva-Is it environmentally sustainable? Algal Res 12:513–521. https://doi.org/10.1016/j.algal.2015.10.015
Guevara G, Lopez MC, Alonso S, Perera J, Navarro-Llorens JM (2019) New insights into the genome of Rhodococcus ruber strain Chol-4. BMC Genomics 20:332. https://doi.org/10.1186/s12864-019-5677-2
Hanway JJ, Heidel H (1952) Soil analysis methods as used in Iowa state college soil testing laboratory. Iowa Agric 57:1–31
Harms H, Koops HP, Wehrmann H (1976) An ammonia-oxidizing bacterium, Nitrosovibrio tenuis nov. gen. nov. sp. Arch Microbiol 108:105–111. https://doi.org/10.1007/BF00425099
Hezayen FF, Rehm BH, Tindall BJ, Steinbüchel A (2001) Transfer of Natrialba asiatica B1T to Natrialba taiwanensis sp. nov. and description of Natrialba aegyptiaca sp. nov., a novel extremely halophilic, aerobic, non-pigmented member of the Archaea from Egypt that produces extracellular poly (glutamic acid). Int J Syst Evol Microbiol 51:1133–1142. https://doi.org/10.1099/00207713-51-3-1133
Jebeli MA, Maleki A, Amoozegar MA, Kalantar E, Izanloo H, Gharibi F (2017) Bacillus flexus strain As-12, a new arsenic transformer bacterium isolated from contaminated water resources. Chemosphere 169:636–641. https://doi.org/10.1016/j.chemosphere.2016.11.129
Jin JJ, Keith PJ, Cummins NW, Kane SV, Pritt BS, Sanchez JL (2017) Lysinibacillus massiliensis panniculitis masquerading as erythema nodosum: a case report. Open Forum Infect Dis. https://doi.org/10.1093/ofid/ofx072
Karthikeyan K, Shanmugam M (2017) The effect of potassium-rich biostimulant from seaweed Kappaphycus alvarezii on yield and quality of cane and cane juice of sugarcane var. Co 86032 under plantation and ratoon crops. J Appl Phycol 29:3245–3252. https://doi.org/10.1007/s10811-017-1211-6
Kleber HP (1997) Bacterial carnitine metabolism. FEMS Microbiol Lett 147:1–9. https://doi.org/10.1111/j.1574-6968.1997.tb10212.x
Kumar R, Trivedi K, Vijay Anand K, Ghosh A (2019) Science behind biostimulant action of seaweed extract on growth and crop yield: Insights into transcriptional changes in roots of maize treated with Kappaphycus alvarezii seaweed extract under soil moisture stressed conditions. J Appl Phycol 32:599–613. https://doi.org/10.1007/s10811-019-01938-y
Layek J, Das A, Ramkrushna GI, Trivedi K, Yesuraj D, Chandramohan M, Kubavat D et al (2015) Seaweed sap: a sustainable way to improve productivity of maize in North-East India. Int J Environ Stud 72:305–315. https://doi.org/10.1080/00207233.2015.1010855
Layek J, Das A, Idapuganti RG, Sarkar D, Ghosh A, Zodape ST, Lal R et al (2018) Seaweed extract as organic bio-stimulant improves productivity and quality of rice in eastern Himalayas. J Appl Phycol 30:547–558. https://doi.org/10.1007/s10811-017-1225-0
Lee JC, Li WJ, Xu LH, Jiang CL, Kim CJ (2008) Lentibacillus salis sp. nov., a moderately halophilic bacterium isolated from a salt lake. Int J Syst Evol Microbiol 58:1838–1843. https://doi.org/10.1099/ijs.0.65545-0
Lekota KE, Bezuidt OKI, Mafofo J, Rees J, Muchadeyi FC, Madoroba E, Van Heerden H (2018) Whole genome sequencing and identification of Bacillus endophyticus and B. anthracis isolated from anthrax outbreaks in South Africa. BMC Microbiol 18:67. https://doi.org/10.1186/s12866-018-1205-9
Lennon JT, Aanderud ZT, Lehmkuhl BK, Schoolmaster DR Jr (2012) Mapping the niche space of soil microorganisms using taxonomy and traits. Ecology 93:1867–1879. https://doi.org/10.1890/11-1745.1
Ling J, Zhu R, Laborda P, Jiang T, Jia Y, Zhao Y, Liu F (2019) LbDSF, the Lysobacter brunescens quorum sensing system diffusible signalling factor, regulates anti-xanthomonas XSAC biosynthesis, colony morphology, and surface motility. Front Microbiol 10:1230. https://doi.org/10.3389/fmicb.2019.01230
Lozupone CA, Hamady M, Kelley ST, Knight R (2007) Quantitative and qualitative β diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microbiol 73(5):1576–1585. https://doi.org/10.1128/AEM.01996-06
Lozupone CA, Stombaugh J, Gonzalez A, Ackermann G, Wendel D, Vázquez-Baeza Y, Jansson JK et al. (2013) Meta-analyses of studies of the human microbiota. Genome Res 23:1704–1714. http://www.genome.org/cgi/doi/http://www.genome.org/cgi/doi.org/10.1101/gr.151803.112
Maynard D, Kalra Y, Crumbaugh J (2006) Soil sampling and methods of analysis. In: Carter M, Gregorich E (eds) 2nd ed. CRC Press, Taylor & Francis Group
Mohamed ZA, Al-Shehri AM (2015) Biodiversity and toxin production of cyanobacteria in mangrove swamps in the Red Sea off the southern coast of Saudi Arabia. Bot Mar 58:23–34. https://doi.org/10.1515/bot-2014-0055
Mondal D, Ghosh A, Prasad K, Singh S, Bhatt N, Zodape ST, Chaudhary JP et al (2015) Elimination of gibberellin from Kappaphycus alvarezii seaweed sap foliar spray enhances corn stover production without compromising the grain yield advantage. Plant Growth Regul 75:657–666. https://doi.org/10.1007/s10725-014-9967-z
Nelson DW, Sommers L (1982) Total carbon, organic carbon, and organic matter 1. Methods soil Anal. Part 2. Chem. Microbiol Prop 539–579
Olsen S, Sommers L (1982) Phosphorus. In: Page A (ed) Methods of soil analysis, part 2: chemical and microbial properties. Soil Science Society of America, Madison, pp 421–422
Orr IG, Hadar Y, Sivan A (2004) Colonization, biofilm formation and biodegradation of polyethylene by a strain of Rhodococcus ruber. Appl Microbiol Biotechnol 65:97–104. https://doi.org/10.1007/s00253-004-1584-8
Oyaert M, De Baere T, Breyne J, De Laere E, Mariën S, Waets P, Laffut W (2013) First case of Pseudoclavibacter bifida bacteremia in an immunocompromised host with chronic obstructive pulmonary disease (COPD). J Clin Microbiol 51:1973–1976. https://doi.org/10.1128/JCM.00138-13
Pang Z, Chen J, Wang T, Gao C, Li Z, Guo L, Xu J, Cheng Y (2021) Linking plant secondary metabolites and plant microbiomes: a review. Front Plant Sci 12:300. https://doi.org/10.3389/fpls.2021.621276
Patel K, Agarwal P, Agarwal PK (2018) Kappaphycus alvarezii sap mitigates abiotic-induced stress in Triticum durum by modulating metabolic coordination and improves growth and yield. J Appl Phycol 30:2659–2673. https://doi.org/10.1007/s10811-018-1423-4
Pradella S, Hans A, Spröer C, Reichenbach H, Gerth K, Beyer S (2002) Characterisation, genome size and genetic manipulation of the myxobacterium Sorangium cellulosum So ce56. Arch Microbiol 178:484–492. https://doi.org/10.1007/s00203-002-0479-2
Pramanick B, Brahmachari K, Ghosh A, Zodape ST (2014) Effect of seaweed saps on growth and yield improvement of transplanted rice in old alluvial soil of West Bengal. Bangladesh J Bot 43:53–58. https://doi.org/10.3329/bjb.v43i1.19746
Pramanick B, Brahmachari K, Ghosh A, Zodape ST (2016) Effect of seaweed saps derived from two marine algae Kappaphycus and Gracilaria on growth and yield improvement of blackgram. Indian J Geo-Mar Sci http://nopr.niscair.res.in/handle/123456789/35111
Pramanick B, Brahmachari K, Mahapatra BS, Ghosh A, Ghosh D, Kar S (2017) Growth, yield and quality improvement of potato tubers through the application of seaweed sap derived from the marine alga Kappaphycus alvarezii. J Appl Phycol 29:3253–3260. https://doi.org/10.1007/s10811-017-1189-0
Quan M, Liang J (2017) The influences of four types of soil on the growth, physiological and biochemical characteristics of Lycoris aurea (L’Her). Herb Sci Rep 7:43284. https://doi.org/10.1038/srep43284
Raats D, Halpern M (2007) Oceanobacillus chironomi sp. nov., a halotolerant and facultatively alkaliphilic species isolated from a chironomid egg mass. Int J Syst Evol Microbiol 57:255–259. https://doi.org/10.1099/ijs.0.64502-0
Rangani J, Parida AK, Panda A, Kumari A (2016) Coordinated changes in antioxidative enzymes protect the photosynthetic machinery from salinity induced oxidative damage and confer salt tolerance in an extreme halophyte Salvadora persica L. Front Plant Science 7:50. https://doi.org/10.3389/fpls.2016.00050
Rathore SS, Chaudhary DR, Boricha GN, Ghosh A, Bhatt BP, Zodape ST, Patolia JS (2009) Effect of seaweed extract on the growth, yield and nutrient uptake of soybean (Glycine max) under rainfed conditions. S Afr J Bot 75:351–355. https://doi.org/10.1016/j.sajb.2008.10.009
Raverkar KP, Pareek N, Chandra R, Chauhan S, Zodape ST, Ghosh A (2016) Impact of foliar application of seaweed saps on yield, nodulation and nutritional quality in green gram (Vigna radiata L). Legum Res 39:315–318. https://doi.org/10.18805/lr.v39i2.9535
Rouphael Y, Giordano M, Cardarelli M, Cozzolino E, Mori M, Kyriacou M, Bonini P et al (2018) Plant-and seaweed-based extracts increase yield but differentially modulate nutritional quality of greenhouse spinach through biostimulant action. Agronomy 8:126. https://doi.org/10.3390/agronomy8070126
Ruan Y, Taherzadeh MJ, Kong D, Lu H, Zhao H, Xu X, Liu Y et al (2020) Nitrogen removal performance and metabolic pathways analysis of a novel aerobic denitrifying halotolerant Pseudomonas balearica strain RAD-17. Microorganisms 8:72. https://doi.org/10.3390/microorganisms8010072
Sameera B, Prakash HS, Nalini MS (2018) Indole acetic acid production by the actinomycetes of coffee plantation soils of Western Ghats. Int J Curr Res 10:74482–74487. https://doi.org/10.24941/ijcr.32796.10.2018
Santos-Medellin C, Edwards J, Liechty Z, Nguyen B, Sundaresan V (2017) Drought stress results in a compartment-specific restructuring of the rice root-associated microbiomes. Mbio 8:e00764-e817. https://doi.org/10.1128/mBio.00764-17
Schnürer J, Rosswall T (1982) Fluorescein diacetate hydrolysis as a measure of total microbial activity in soil and litter. Appl Environ Microbiol 43:1256–1261. https://doi.org/10.1128/aem.43.6.1256-1261.1982
Sharma L, Banerjee M, Malik GC, Vijayanand KG, Zodape ST, Ghosh A (2017) Sustainable agro-technology for enhancement of rice production in the red and lateritic soils using seaweed based biostimulants. J Clean Prod 149:968–975. https://doi.org/10.1016/j.jclepro.2017.02.153
Shirsalimian MS, Amoozegar MA, Sepahy AA, Kalantar SM, Dabbagh R (2017) Isolation of extremely halophilic Archaea from a saline river in the Lut Desert of Iran, moderately resistant to desiccation and gamma radiation. Microbiology 86:403–411. https://doi.org/10.1134/S0026261717030158
Singh G, Mukerji KG (2006) Root exudates as determinant of rhizospheric microbial biodiversity. In: Mukerji KG, Manoharachary C, Singh J (eds) Microbial activity in the rhizosphere. Soil biology, vol 7. Springer, Berlin Heidelberg. https://doi.org/10.1007/3-540-29420-1_3
Singh S, Prakash A, Chakraborty NR, Wheeler C, Agarwal PK, Ghosh A (2016a) Trait selection by path and principal component analysis in Jatropha curcas for enhanced oil yield. Ind Crops Prod 86:173–179. https://doi.org/10.1016/j.indcrop.2016.03.047
Singh S, Singh MK, Pal SK, Trivedi K, Yesuraj D, Singh CS, Vijayanand KGV et al (2016b) Sustainable enhancement in yield and quality of rain-fed maize through Gracilaria edulis and Kappaphycus alvarezii seaweed sap. J Appl Phycol 28:2099–2112. https://doi.org/10.1007/s10811-015-0680-8
Singh I, Vijayanand KG, Solomon S, Shukla SK, Rai R, Zodape ST, Ghosh A (2018) Can we not mitigate climate change using seaweed based biostimulant: a case study with sugarcane cultivation in India. J Clean Prod 204:992–1003. https://doi.org/10.1016/j.jclepro.2018.09.070
Song F, Han X, Zhu X, Herbert SJ (2012) Response to water stress of soil enzymes and root exudates from drought and non-drought tolerant corn hybrids at different growth stages. Can J Soil Sci 92:501–507. https://doi.org/10.4141/cjss2010-057
Tabatabai M (1982) Soil enzymes. In: Page AL (ed) Methods of soil analysis. Part 2. Chemical and microbiological properties. American Society of Agronomy Soil Science Society of America, pp 903–947
Tamura T, Hayakawa M, Nonomura H, Yokota A, Hatano K (1995) Four New Species of the Genus Actinokineospora: Actinokineospora inagensis sp. nov., Actinokineospora globicatena sp. nov., Actinokineospora terrae sp. nov., and Actinokineospora diospyrosa sp. nov. Int J Syst Evol 45:371–378. https://doi.org/10.1099/00207713-45-2-371
Tamura T, Hayakawa M, Hatano K (2001) A new genus of the order Actinomycetales, Virgosporangium gen. nov., with descriptions of Virgosporangium ochraceum sp. nov. and Virgosporangium aurantiacum sp. nov. Int J Syst Evol Microbiol 51:1809–1816. https://doi.org/10.1099/00207713-51-5-1809
Tanaka K, Stackebrandt E, Tohyama S, Eguchi T (2000) Desulfovirga adipica gen. nov., sp. nov., an adipate-degrading, gram-negative, sulfate-reducing bacterium. Int J Syst Evol Microbiol 50:639–644. https://doi.org/10.1099/00207713-50-2-639
Tatton MJ, Archer DB, Powell GE, Parker ML (1989) Methanogenesis from ethanol by defined mixed continuous cultures. Appl Environ Microbiol 55:440–445. https://doi.org/10.1128/aem.55.2.440-445.1989
Tiago I, Pires C, Mendes V, Morais PV, Da Costa MS, Verissimo A (2006) Bacillus foraminis sp. nov., isolated from a non-saline alkaline groundwater. Int J Syst Evol Microbiol 56:2571–2574. https://doi.org/10.1099/ijs.0.64281-0
Tilman D, Cassman KG, Matson PA, Naylor R, Polasky S (2002) Agricultural sustainability and intensive production practices. Nature. https://doi.org/10.1038/nature01014
Trivedi K, Vijayanand KG, Kubavat D, Kumar R (2017) Crop stage selection is vital to elicit optimal response of maize to seaweed bio-stimulant application. J Appl Phycol 29:2135–2144. https://doi.org/10.1007/s10811-017-1118-2
Trivedi K, Vijayanand KG, Kubavat D, Patidar R, Ghosh A (2018a) Drought alleviatory potential of Kappaphycus seaweed extract and the role of the quaternary ammonium compounds as its constituents towards imparting drought tolerance in Zea mays L. J Appl Phycol 30:2001–2015. https://doi.org/10.1007/s10811-017-1375-0
Trivedi K, Vijayanand KG, Vaghela P, Ghosh A (2018b) Differential growth, yield and biochemical responses of maize to the exogenous application of Kappaphycus alvarezii seaweed extract, at grain-filling stage under normal and drought conditions. Algal Res 35:236–244. https://doi.org/10.1016/j.algal.2018.08.027
Wang J, Liu B, Liu G, Pan Z, Xiao R, Chen M, Chen D (2016) Draft genome sequence of Bacillus humi LMG 22167T (DSM 16318), an endospore-forming bacterium isolated from soil. Genome Announc. https://doi.org/10.1128/genomeA.01692-15
Wells B, Burnum AL, Armstrong J, Sanchez S, Stanton JB, Camus MS (2018) Actinomadura vinacea isolated from a nonhealing cutaneous wound in a cat. Vet Clin Pathol 47:638–642. https://doi.org/10.1111/vcp.12659
Whalen JK, Sampedro L (2009) Soil Ecology and Management. CABI. https://doi.org/10.1079/9781845935634.0000
Xie CH, Yokota A (2005) Pleomorphomonas oryzae gen. nov., sp. nov., a nitrogen-fixing bacterium isolated from paddy soil of Oryza sativa. Int J Syst Evol Microbiol 55:1233–1237. https://doi.org/10.1099/ijs.0.63406-0
Yamamura S, Yamashita M, Fujimoto N, Kuroda M, Kashiwa M, Sei K, Fujita M et al (2007) Bacillus selenatarsenatis sp. nov., a selenate-and arsenate-reducing bacterium isolated from the effluent drain of a glass-manufacturing plant. Int J Syst Evol Microbiol 57:1060–1064. https://doi.org/10.1099/ijs.0.64667-0
Yumoto I, Iwata H, Sawabe T, Ueno K, Ichise N, Matsuyama H, Okuyama H et al (1999) Characterization of a facultatively psychrophilic bacterium, Vibrio rumoiensis sp. nov., that exhibits high catalase activity. Appl Environ Microbiol 65:67–72. https://doi.org/10.1128/AEM.65.1.67-72.1999
Zhang K, Tang Y, Zhang L, Dai J, Wang Y, Luo X, Liu M et al (2009) Parasegetibacter luojiensis gen. nov., sp. nov., a member of the phylum Bacteroidetes isolated from a forest soil. Int J Syst Evol Microbiol 59:3058–3062. https://doi.org/10.1099/ijs.0.008763-0
Zodape ST, Gupta A, Bhandari SC, Rawat US, Chaudhary DR, Eswaran K, Chikara J (2011) Foliar application of seaweed sap as biostimulant for enhancement of yield and quality of tomato (Lycopersicon esculentum Mill). J Sci Ind Res 70:215–291. http://nopr.niscair.res.in/handle/123456789/11089
Acknowledgements
KT and DK would like to thank CSIR and AcSIR-CSMCRI for the award of senior research fellowship (SRF) and enrollment in a Ph.D. program. The authors also wish to thank CSMCRI MARS Mandapam Camp, Tamil Nadu, India, especially Dr. K Eswaran and Dr. Vaibhav Mantri for useful discussion and support. Dr Shruti Chatterjee is acknowledged for providing bacterial strains. SciGenom Labs Private Limited, Cochin, Kerala is duly acknowledged for help in sequencing analysis. This manuscript bears CSIR-CSMCRI communication no. 132/2019.
Funding
This work was supported by the council of scientific and industrial research (CSIR) under the project MLP 0016.
Author information
Authors and Affiliations
Contributions
KT performed experimental work and contributed to data acquisition, its analysis and interpretation. RK, GB and DK contributed during data acquisition and drafting of the article. KGV contributed to data acquisition and drafting of manuscript and reviewed the manuscript critically for important intellectual content. AG designed the experiment, coordinated the work, interpreted data and critically reviewed the manuscript. All the authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflicts of interest to declare that are relevant to the content of this article.
Additional information
Communicated by Erko Stackebrandt.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Trivedi, K., Kumar, R., Vijay Anand, K.G. et al. Structural and functional changes in soil bacterial communities by drifting spray application of a commercial red seaweed extract as revealed by metagenomics. Arch Microbiol 204, 72 (2022). https://doi.org/10.1007/s00203-021-02644-5
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00203-021-02644-5