Abstract
Saponins are kinds of antifungal compounds produced by Panax notoginseng to resist invasion by pathogens. Ilyonectria mors-panacis G3B was the dominant pathogen inducing root rot of P. notoginseng, and the abilities to detoxify saponins were the key to infect P. notoginseng successfully. To research the molecular mechanisms of detoxifying saponins in I. mors-panacis G3B, we used high-throughput RNA-Seq to identify 557 and 1519 differential expression genes (DEGs) in I. mors-panacis G3B with saponins treatments for 4H (Hours) and 12H (Hours) compared with no saponins treatments, respectively. Among these DEGs, we found 93 genes which were simultaneously highly expressed in I. mors-panacis G3B with saponins treatments for 4H and 12H, they mainly belong to genes encoding transporters, glycoside hydrolases, oxidation–reduction enzymes, transcription factors and so on. In addition, there were 21 putative PHI (Pathogen–Host Interaction) genes out of those 93 up-regulated genes. In this report, we analyzed virulence-associated genes in I. mors-panacis G3B which may be related to detoxifying saponins to infect P. notoginseng successfully. They provided an excellent starting point for in-depth study on pathogenicity of I. mors-panacis G3B and developed appropriate root rot disease management strategies in the future.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Panax notoginseng (Burkill) F.H. Chen (P. notoginseng) is a kind of traditional Chinese medicinal herb mainly used for treatment of a variety of diseases, such as ischemic cardiovascular diseases and so on. P. notoginseng which belongs to the Panax genus and Araliaceae has been cultivated for more than 400 years in china since the time of Shizhen Li who compiled “Compendium of Materia Medica”. Currently, it was mainly cultivated artificially in the mountain area with altitude between 1200 and 2000 m of Wenshan prefecture, Yunnan province, China (Guo et al. 2010). Although there had been reports on wild species of P. notoginseng, no successful case had been described and only some closely related wild species or variety had been found (Yan et al. 2006). P. notoginseng is a perennial medicinal plant that grows in the shade and needs 4–6 years to produce mature roots and accumulate of bioactive compounds such as saponins. The humid production environment and prolonged period of growth expose the roots to potential infection by different pathogens, which severely restrict the production of P. notoginseng, especially in land-limited mountain area.
P. notoginseng is vulnerable to be attacked by soil microbes including fungi, bacteria and nematodes because of its long-term cultivation and shady environment. Fungi dominate with increasing years of planting, more than 70 genera were found in the rhizosphere soil of P. notoginseng cultivated in Wenshan prefecture, and 20 species have been identified (Wang et al. 2003). Pathogenic fungi cause serious diseases, such as black spot, circular spot, gray mold, root rot and so on. Among them, root rot is the most common and severe disease, resulting in dramatic crop losses up to 10–20%, or more than 70% (Wu et al. 2015; Sun et al. 2004). Root rot typically appears at the tip of the taproot destroying the fibrous roots and attacks toward the crown, the core of the root eventually disintegrates and remains hollow, which is also known as disappearing root rot (Rahman and Punja 2005). Thus, identification of the dominant pathogens of the root rot and clarification their pathogenesis are prerequisites for effective control to maintain the sustainable cultivation of P. notoginseng.
Root rot is an ubiquitous disease worldwide, occurring in multiple plants with varied pathogens. And Cylindrocarpon destructans is a kind of soilborne pathogenic fungi which can cause severe root rot in many hosts including P. ginseng. They can be divided into weak and aggressive isolates. However, aggressive isolates cause severe root rot disease especially to P. gingseng with limited pathogenicity in other hosts, and were therefore named C. destructans f. sp. Panacis (Seifert et al. 2003). Then, C. destructans was reclassified as Ilyonectria radicicola (Chaverri et al. 2011). According to multigene molecular analysis especially histone H3 (HIS H3), I. radicicola isolates appeared a polyphyletic relationship and each group of isolates is considered to be a different species (Lombard et al. 2013; Cabral et al. 2012). And C. destructans f. sp. panacis was genetically distinct from the other isolates and clustered in a distinct group named as Ilyonectria mors-panacis (Seifert et al. 2003). On these bases, Mi used culture-dependent and molecular methods to investigate the fungal communities and identify the dominant pathogen of G3B inducing root rot of P. notoginseng based on in vitro and in vivo pathogenicity. G3B is phylogenetically and phenotypically similar to I. mors-panacis, so named for Ilyonectria mors-panacis G3B (Mi et al. 2017). And Zhu reported available genome sequence of I. mors-panacis G3B and its annotation, the annotation analysis predicted 120 virulence genes defined by the Database of Virulence Factors in Fungal Pathogens including 28 cell wall degrading enzymes, 18 antioxidant systems, 7 effectors and so on. This further proved that I. mors-panacis G3B is the dominant agreesive pathogen causing root rot of P. notoginseng (Zhu et al. 2019). Therefore, understanding the genetic properties and pathogenicity of I. mors-panacis will provide theortical supports for inhibiting root rot to maintain sustainable cultivation of P. notoginseng.
Ilyonectria mors-panacis has complicated pathogenicity processes, they mainly include the following steps: when spore or mycelium adheres to the root surface, I. mors-panacis rapidly produces high quantities of hydrolytic enzymes, such as cellulase and pectina mors-panacis, allowing rapid invasion of the epidermal layer and fast extension of the inoculum to the cortical and inner tissues; And the ginseng plant starts to secret ginsenosides or phenolic compounds to resist the invasion by pathogens. In response to them, I. mors-panacis further produces enzymes that can degrade ginsenosides or phenolic compounds, such as glycosidases and polyphenoloxidases. At the same time, I. mors-panacis sequesters iron from the ginseng plant to support its growth using siderophores; once the cell wall components of ginseng plant break down, its defense response declines and I. mors-panacis propagates quickly and then root rotting symptoms are established (Morrissey et al. 2000; Rahman and Punja 2005; Ivanov and Bernards 2012; Farh et al. 2018). During the pathogenicity processes, ginsenosides secreted by ginseng plant have been shown to possess chemical defenses against fungi and therefore act as phytoanticipins (Nicol et al. 2002). However, the antifungal action of ginsenosides is not effective against all potential ginseng pathogens. Oppositely, they have been shown to stimulate the in vitro growth of some pathogens inducing root rot of ginseng plants and this may result from the ability of these pathogens to metabolize ginsenosides via extracellular glycosidases (Andreea Neculai et al. 2009; LF and MA 2006). In addition, relevant study has shown that the highly aggressive species of pathogens inducing root rot disease produces much more hydrolytic enzyme, the oxidative enzyme and polyphenol oxidase than weakly aggressive, which destroy the plant defensive barriers (Rahman and Punja 2005). However, the detail involvements of hydrolytic enzymes in detoxifying ginsenosides and more related pathogenesis have yet been studied.
Therefore, the goal of the present study is to analyze the genes encoding the saponins degrading enzymes from I. mors-panacis G3B. To achieve this objective, we built the transcriptome sequencing platform for high-throughput prediction of all associated genes. This study not only revealed candidate genes for further functional research on pathogenicity of I. mors-pannacis but also provided theoretical supports for inhibiting root rot and alleviating replant failure of ginseng plants.
Materials and methods
Fungal strains and growth conditions
Ilyonectria mors-panacis G3B used in the study was first isolated from the rhizosphere soil of diseased P. notoginseng cultivated in Wenshan, Yunnan Province, China (Mi et al. 2017). The strain was cultured onto a PDA (Potato Dextrose-Agar) plate at 22 ℃ for 18 days.
Assessment of the tolerance of Ilyonectria mors-panacis G3B to saponins
The susceptibility of I. mors-panacis G3B to saponins was evaluated by estimating the growth diameters of colonies which were inoculated in PDA agars and those supplemented with Sanqi total Saponins (SAPs) (Solarbio). 5 µL conidial suspension (1 × 107 conidia mL−1) was, respectively, applied to the center of PDA agar plates (90 mm in diameter) and those containing SAPs at the concentration of 500 ppm. Inoculated plates were incubated at 22 ℃ and colonies diameters were measured daily from 3 days post inoculation until the colonies ceased growing. Three samples per treatment were used as replicates and the experiment was conducted three times.
Assessment of the capacities of Ilyonectria mors-panacis G3B to degrade saponins
Conidial suspensions of I. mors-panacis G3B were inoculated in the PDA liquid medium supplemented with proper amount of saponins. And non-inoculations of I. mors-panacis G3B in the PDA liquid medium containing equivalent saponins were negative controls. According to the standard curves (concentration-peak area) of Rg1 and Rb1 analyzed by HPLC, we assayed the concentrations of Rg1 and Rb1 after 0 day, 3 days and 6–12 days inoculation in the PDA culture solution, respectively.
I. mors-panacis G3B incubation with saponins and RNA extraction
The conidia of I. mors-panacis G3B were inoculated in sabouraud dextrose broth at a final concentration of 1 × 106 conidia mL−1 and incubated for 36 h at 22℃ with 220 rpm. The mycelium was collected by filtration, and washed three times with sterilized water. Subsequently, 0.5 g mycelia were inoculated into 50 mL basal salt solution (M100 medium with glucose excluded) and equivalent basal salt solution added with saponins as sole carbon and nitrogen sources, respectively, then incubated at 22℃ with 220 rpm for 4H and 12H. The mycelia of I. mors-panacis G3B at each time point were collected and rinsed with distilled water and then immediately frozen in liquid nitrogen until RNA extraction. The mycelium harvested from I. mors-panacis G3B incubated with saponins-free basal salt solution served as controls (4H and 12H). The total RNA was extracted using TRIzol Reagent in accordance with manufacturer’s protocols and then treated with RNase-free DNase to eliminate genomic DNA contamination. The total RNA was quantified on the Thermo Scientific NanoDrop 2000 spectrophotometer and the Agilent 2100 Bioanalyzer.
cDNA library construction and sequencing
The mRNA was purified and isolated by treating total RNA with Magnetic Oligo (dT) beads. Then, the purified mRNA was sheared to approximately 200 bp fragments prior to cDNA synthesis. Short fragments were purified and ligated to sequencing adapters. Fragments with suitable sizes on the basis of agarose gel electrophoresis were selected as templates for PCR amplification to isolate and purify the cDNA fragments for sequencing. Construction of libraries (Illumina Truseq™ RNA sample prep Kit) and sequencing with the Illumina HiSeq 2000 platform were performed by Frasergen (Shanghai, China). The quality of raw RNA-Seq reads was filtered using the following criteria: (1) reads including adapter sequencing or empty adapter were filtered; (2) reads for which Ns comprised more than 10% of the total length were discarded; (3) reads with low-quality bases (< Q20) were filtered.
RNA-Seq reads mapping and annotation
Hisat2 v 2.1.0 was used to map the RNA-Seq reads with the reference genome for subsequent analysis. The mapped reads were subjected to de novo transcriptome assembly using trinity assembly software to obtain high-quality transcript sequence. Then, the assembled sequences were used for a homology search against the NR, String, Swissport and KEGG database by NCBI-Blastx Version 2.2.25 with an E-value of 10–5.
Gene expression analysis and DEGs validation
Reads that aligned uniquely to the reference sequence were used for gene expression quantification that were measured and normalized as the fragments per kilobase of exon per million fragments mapped (FPKM), which is similar to reads per kilobase of exon per million mapped reads (Marioni et al. 2008). Differential expression analysis was performed with edgeR v 3.24 software using the test of fold change (|log2FPKM|> 1) and false discovery rate (fdr < 0.05) to estimate the level of differential gene expression by each sample under different induction conditions (Benjamini et al. 2001).
Analysis virulence-associated genes during saponins metabolism
To identify potential pathogenicity and virulence genes, whole genome blast searches were conducted against protein sequences in the Pathogen–Host Interaction database (PHI database) (version 3.2, http://www.phi-base.org/) (E < 1 × 10–5). Further, we found the partial virulence-associated genes identified from the PHI database among differential expression genes at 4H and 12H during saponins metabolism.
Results
The tolerance of Ilyonectria mors-panacis G3B to saponins
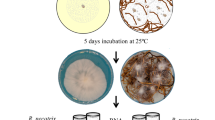
P. notoginseng can secret secondary metabolites such as saponins that used for anti-microbes. Therefore, we assayed the sensitivity to saponins of I. mors-panacis G3B as dominant pathogen inducing root rot of P. notoginseng. Under saponins stress [PDA supplemented with 500 ppm Sanqi total Saponins (SAPs)], I. mors-panacis G3B grew significantly faster and produced bigger isolated colonies than control (Inoculation on PDA without SAPs) (P < 0.05) (Fig. 1). They suggested that I. mors-panacis G3B could degrade saponins and use them as carbon sources, and this might also be the main factor for I. mors-panacis G3B to infect the P. notoginseng successfully.
The tolerance of I. mors-panacis G3B to Sanqi total Saponins (SAPs). (A) Growth curves of I. mors-panacis G3B on PDA plates without SAPs and PDA plates supplemented with SAPs (500 ppm). The star indicates that the growth of I. mors-panacis G3B on the PDA plates supplemented with SAPs was significantly faster than that on the PDA plates without SAPs (P < 0.05). (B) Morphology of colonies of I. mors-panacis G3B. Colony pictures were taken at 15 days post inoculation by applying 5 μL of a conidial suspension (1 × 107 conidia mL−1) inoculated on the PDA without SAPs and PDA supplemented with SAPs (500 ppm). Scale bars represent 10 mm
The capacity of Ilyonectria mors-panacis G3B to detoxify saponins
To verify the degradation effect on saponins of I. mors-panacis G3B directly, we assayed its abilities to degrade the Rg1 and Rb1. Compared with CK, the concentration of Rg1 and Rb1 in the culture solution inoculation with I. mors-panacis G3B was significantly decreased at 3 days and 6–12 days post inoculation, respectively (Fig. 2). They indicated that I. mors-panacis G3B could degrade different kinds of saponins, such as Rg1 and Rb1, and exhibiting different degradation abilities.
The concentration of Rg1 (A) and Rb1 (B) in the culture solution at different time post inoculation with I. mors-panacis G3B or without inoculation (CK). The stars indicate that the concentration of Rg1 and Rb1 in the culture solution inoculation with I. mors-panacis G3B was significantly decreased compared with CK (P < 0.05). The experiments were repeated three times with three replicates per repeat
Overview of RNA-Seq analysis of I. mors-panacis G3B
In general, saponins exhibit antifungal activities against soil-borne fungi. However, the pathogenic fungi can produce saponins detoxifying enzymes to decrease the fungi toxicity (Watanabe et al. 2004; Yousef and Bernards, 2006). To find the genes which may be related to detoxifying saponins, the transcriptomes of I. mors-panacis G3B which were treated with saponins for 4H and 12H were profiled by Illumina HiSeq 2000 RNA-Seq (free-saponins treatment as negative control), two biological replicates and 8 datasets were established. Approximately 30 million 200 bp paired-end reads were generated, the majority of the reads (~ 94%) were mapped to the I. mors-panacis G3B draft reference genome sequence which had been available from the GenBank under the accession number PPHJ00000000.1 (BioProject: PRJNA431033) and more than 70% unique mapped reads appeared, indicating the high abundance and excellent quality of the sequencing data (Table 1). In this study, one gene was considered to be expressed when its fragments per kilobase per million fragments (FPKM) were greater than or equal to 1 and genes with FPKM between 8 and 32 represented a majority (Fig. 3).
DEGs identification
To identify the putative signal transduction and metabolic pathways involved in detoxifying saponins in I. mors-panacis G3B, we used RNA-Seq to compare the set of differentially expressed genes (DEGs) between saponins treatments and free-saponins treatments for 4H and 12H, respectively. In this section, ‘up-regulated genes’ are genes with higher expression levels when saponins treatment, and ‘down-regulated genes’ are those with lower expression levels under the same conditions. When mycelium of I. mors-panacis G3B grown for 4H and 12H, the number of DEGs was 557 (247 up-regulated and 310 down-regulated) and 1519 (683 up-regulated and 836 down-regulated), respectively. In addition, 343 genes including 93 up-regulated genes and 249 down-regulated genes presented the same expression pattern in the mycelium of I. mors-panacis G3B with saponins treatments compared with free-saponins treatments for 4H and 12H (Fig. 4).
Transcriptomic analysis of I. mors-panacis G3B during saponins catabolic process. (A) DEGs of mycelium incubated with saponins compared with no saponins treatments for 4H and 12H, respectively. (B) Venn diagram showing the distribution of shared DEGs of mycelium incubated for different time courses (4H and 12H). Shown in the parentheses are the number of upregulated genes (red) and downregulated genes (green) in I. mors-panacis G3B with saponins treatments for 4H and 12H compared with control (without saponins treatments)
Core genes encoding saponins detoxifying enzymes
Saponins are kinds of glycosides whose aglycones are triterpenes or spiral steranes, they are composed of saponin units and sugars, such as glucose, galactose, rhamnose, arabinose, glucuronic acid, galacturonic acid and so on. And the genes involved in their metabolism are unknown. According to the above RNA-Seq analysis, if a DEG in the I. mors-panacis G3B with saponins treatments had a similar expression pattern between 4 and 12H post inoculation, it may be involved in signal transduction and metabolic pathway of saponins. Therefore, we mainly focused on the above 93 up-regulated genes simultaneously present in the I. mors-panacis G3B treated with saponins for 4H and 12H, they mainly included several kinds of transporters, glycoside hydrolases, oxidation–reduction enzymes, transcription factors and so on (Fig. 5). We speculated that they are responsible for transporting and metabolizing saponins, making I. mors-panacis G3B resist the antimicrobial activity of saponins and infect P. notoginseng successfully.
Expression patterns of some transporters (A), hydrolases (B), transcription factors (TFs) (C), oxidoreductases (D) and ATPases (E) in I. mors-panacis G3B. C represent controls, I. mors-panacis G3B were inoculated in the basal salt solution without saponins for 4H and 12H; S represent samples, I. mors-panacis G3B were inoculated in the basal salt solution with saponins for 4H and 12H
Virulence-associated genes
To find potential virulence-associated genes, the whole genome blast analysis was conducted against the Pathogen–Host Interaction (PHI) gene database, a collection of experimentally verified pathogenicity, virulence and effector genes from fungi, oomycetes and bacteria (Winnenburg et al. 2007). And we identified 2298 putative PHI genes in I. mors-panacis G3B (12.5% of its genes). We processed on the assumption that the proof of pathogenicity or virulence of a gene in one fungus may also suggest a pathogenicity or virulence in other fungi (Baldwin et al. 2006). Therefore, the 2298 putative PHI genes may be involved in pathogenicity in I. mors-panacis G3B, and we found 21 genes which were simultaneously highly expressed in I. mors-panacis G3B treated with saponins for 4H and 12H among them (Table 2). They may be pathogenicity determinants and involved in detoxifying saponins as antimicrobial compounds.
Discussion
Ginsenosides are antifungal compounds that are thought to be secreted by ginseng plants to defense infection by soilborne fungi (Augustin et al. 2011), their concentrations dramatically increase in adventitious hairy roots of Panax ginseng when attacked by microbes (Liu et al. 2004). However, ginsenosides showed a rapid reduction in roots infected with the aggressive I. mors-panacis isolates (Farh et al. 2017). Relevant study also showed that when the tomato was artificially inoculated with Cladosporium fulvum inducing blight spot, α-tomatine secreted by tomato plants reduced because of β-glucosidase produced by pathogen hydrolyzing it, resulting in accumulation of a less fungi-toxic compound (Okmen et al. 2013). In the current study, we found the growth of I. mors-panacis G3B was significantly increased in media supplemented with saponins, indicating that I. mors-panacis G3B could produce associated saponin-hydrolyzing enzymes to metabolize saponins. Therefore, I. mors-panacis may use a similar mechanism to detoxify ginsenosides, such as Rg1 and Rb1 and so on, and resist antifungal activity to infect the ginseng plants successfully.
Recent improvements in next-generation sequencing technology and bioinformatics now allow the de novo assembly of high-quality eukaryotic genome (Nowrousian et al. 2010; Li et al. 2010). Previously, we used such an approach to provide the first draft sequences of I. mors-panacis G3B inducing root rot of P. notoginseng, and thus serve as an excellent starting point for gaining a broad perspective of issues in P. notoginseng pathology (Zhu et al. 2019). In this study, we used high-throughput RNA-Seq to characterize the transcriptome profile of I. mors-panacis G3B with saponins treatments for different time periods. The induced responses of I. mors-panacis G3B to saponins treatments were characterized to reveal genes involved in the saponins detoxification. Saponins, based on a dammarane carbon skeleton with four trans-oriented rings and side chains that consist various sugar moieties (mono- and disaccharides of glucose, rhamnose, xylose and arabinose) attached through the C-20 and either the C-3 or C-6 positions (Attele et al. 1999). By comprehensive analysis, a total of 93 unigenes were all up-regulated in mycelium harvested from I. mors-panacis G3B treated with saponins for 4H and 12H. The above-related gene encoding proteins mainly belong to transporters, glycoside hydrolases, oxidation–reduction enzymes, transcription factors and therefore they may be involved in putative signal transduction and metabolic pathways to detoxify saponins.
Using the experimentally verified Pathogen–Host Interaction (PHI) gene reference database (Winnenburg et al. 2007), we found that 12.5% of the genes (2298) in the I. mors-panacis G3B genome have significant similarities with genes involved in pathogenicity in other fungi, such as plant pathogens F. graminearum and M.oryzae and even animal pathogens C. albicans, they may be candidate genes controlling pathogenicity. Further, we identified 21 genes which were simultaneously highly expressed in I. mors-panacis G3B treated with saponins for 4H and 12H among the 2298 genes, they may be involved in saponins metabolism for resisting antifungal activity to control pathogenicity.
In conclusion, our study explored the tolerance of I. mors-panancis G3B to saponins, which may intervene in the plant defense mechanism against pathogens infecting ginseng plants. The pathogenicity of I. mors-panacis G3B may mainly depended on detoxification saponins and we analyzed associated virulence genes which may be involved in saponins metabolism. They provide an excellent starting point for in-depth study of biological function of related genes in the further work and gain advanced insights into the pathogenicity of I. mors-panacis causing root rot in ginseng plants.
References
Andreea Neculai M, Ivanov D, Bernards MA (2009) Partial purification and characterization of three ginsenoside-metabolizing β-glucosidases from Pythium irregulare. Phytochemistry 70:1948–1957. https://doi.org/10.1016/j.phytochem.2009.09.007
Attele AS, Wu JA, Yuan CS (1999) Ginseng pharmacology: multiple constituents and multiple actions. Biochem Pharmacol 58:1685–1693. https://doi.org/10.1016/S0006-2952(99)00212-9
Augustin JM, Kuzina V, Andersen SB, Bak S (2011) Molecular activities, biosynthesis and evolution of triterpenoid saponins. Phytochemistry 72:435–457. https://doi.org/10.1016/j.phytochem.2011.01.015
Baldwin TK, Winnenburg R, Urban M, Rawlings C, Koehler J, Hammond-Kosack KE (2006) The pathogen-host interactions database (PHI-base) provides insights into generic and novel themes of pathogenicity. Mol Plant Microbe Interact 19:1451–1462. https://doi.org/10.1094/MPMI-19-1451
Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I (2001) Controlling the false discovery rate in behavior genetics research. Behav Brain Res 125:279–284. https://doi.org/10.1016/S0166-4328(01)00297-2
Cabral A, Groenewald JZ, Rego C, Oliveira H, Crous PW (2012) Cylindrocarpon root rot: multi-gene analysis reveals novel species within the Ilyonectria radicicola species complex. Mycol Prog 11:655–688. https://doi.org/10.1007/s11557-011-0777-7
Chaverri P, Salgado C, Hirooka Y, Rossman AY, Samuels GJ (2011) Delimitation of Neonectria and Cylindrocarpon (Nectriaceae, Hypocreales, Ascomycota) and related genera with Cylindrocarpon-like anamorphs. Stud Mycol 68:57–78. https://doi.org/10.3114/sim.2011.68.03
Farh ME, Kim Y, Singh P, Yang D (2017) Cross Interaction Between Ilyonectria mors-panacis Isolates Infecting Korean Ginseng and Ginseng Saponins in Correlation with Their Pathogenicity. Phytopathology 107:561–569. https://doi.org/10.1094/PHYTO-05-16-0210-R
Farh ME, Kim Y, Kim Y, Yang D (2018) Cylindrocarpon destructans/Ilyonectria radicicola-species complex: causative agent of ginseng root-rot disease and rusty symptoms. J Ginseng Res 42:9–15. https://doi.org/10.1016/j.jgr.2017.01.004
Guo HB, Cui XM, An N, Cai GP (2010) Sanchi ginseng (Panax notoginseng (Burkill) F. H. Chen) in China: distribution, cultivation and variations. Genet Resour Crop Evol 57:453–460. https://doi.org/10.1007/s10722-010-9531-2
Ivanov DA, Bernards MA (2012) Ginsenosidases and the pathogenicity of Pythium irregulare. Phytochemistry 78:44–53. https://doi.org/10.1016/j.phytochem.2012.02.024
LF, Y., and B. MA. (2006) In vitro metabolism of ginsenosides by the ginseng root pathogen Pythium irregulare. Phytochemistry 67:1740–1749. https://doi.org/10.1016/j.phytochem.2005.06.030
Li R, Fan W, Tian G, Zhu H, He L, Cai J, Huang Q, Cai Q, Li B, Bai Y, Zhang Z, Zhang Y, Wang W, Li J, Wei F, Li H, Jian M, Li J, Zhang Z, Nielsen R, Li D, Gu W, Yang Z, Xuan Z, Ryder OA, Leung FC, Zhou Y, Cao J, Sun X, Fu Y, Fang X, Guo X, Wang B, Hou R, Shen F, Mu B, Ni P, Lin R, Qian W, Wang G, Yu C, Nie W, Wang J, Wu Z, Liang H, Min J, Wu Q, Cheng S, Ruan J, Wang M, Shi Z, Wen M, Liu B, Ren X, Zheng H, Dong D, Cook K, Shan G, Zhang H, Kosiol C, Xie X, Lu Z, Zheng H, Li Y, Steiner CC, Lam TT, Lin S, Zhang Q, Li G, Tian J, Gong T, Liu H, Zhang D, Fang L, Ye C, Zhang J, Hu W, Xu A, Ren Y, Zhang G, Bruford MW, Li Q, Ma L, Guo Y, An N, Hu Y, Zheng Y, Shi Y, Li Z, Liu Q, Chen Y, Zhao J, Qu N, Zhao S, Tian F, Wang X, Wang H, Xu L, Liu X, Vinar T et al (2010) The sequence and de novo assembly of the giant panda genome. Nature 463:311–317. https://doi.org/10.1038/nature08696
Liu J, Ding JY, Zhou QY, He L, Wang ZT (2004) Studies on influence of fungal elicitor on hairy root of Panax ginseng biosynthesis ginseng saponin and biomass. China J Chin Materia Med 29:302–305
Lombard L, Bezuidenhout CM, Crous PW (2013) Ilyonectria black foot rot associated with Proteaceae. Australas Plant Pathol 42:337–349. https://doi.org/10.1007/s13313-012-0188-5
Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y (2008) RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res 18:1509–1517
Mi C, Yang R, Rao J, Yang S, Wei F, Li O, Hu X (2017) Unveiling of dominant fungal pathogens associated with rusty root rot of Panax notoginseng based on multiple methods. Plant Dis 101:2046–2052. https://doi.org/10.1094/PDIS-01-17-0135-RE
MORRISSEY, J. P., J. P. WUBBEN, and A. E. OSBOURN. (2000) Stagonospora avenae secretes multiple enzymes that hydrolyze oat leaf saponins. Mol Plant Microbe Interact 13:1041–1052. https://doi.org/10.1094/MPMI.2000.13.10.1041
Nicol RW, Traquair JA, Bernards MA (2002) Ginsenosides as host resistance factors in American ginseng (Panax quinquefolius). Can J Bot 80:557–562. https://doi.org/10.1139/B02-034
Nowrousian M, Stajich JE, Chu M, Engh I, Espagne E, Halliday K, Kamerewerd J, Kempken F, Knab B, Kuo HC, Osiewacz HD, Poggeler S, Read ND, Seiler S, Smith KM, Zickler D, Kuck U, Freitag M (2010) De novo assembly of a 40 Mb eukaryotic genome from short sequence reads: Sordaria macrospora, a model organism for fungal morphogenesis. PLoS Genet 6:e1000891. https://doi.org/10.1371/journal.pgen.1000891
Okmen B, Etalo DW, Joosten MH, Bouwmeester HJ, de Vos RC, Collemare J, de Wit PJ (2013) Detoxification of α-tomatine by Cladosporium fulvum is required for full virulence on tomato. New Phytol 198:1203–1214. https://doi.org/10.1111/nph.12208
Rahman M, Punja ZK (2005) Factors influencing development of root rot on ginseng caused by Cylindrocarpon destructans. Phytopathology 95:1381–1390. https://doi.org/10.1094/PHYTO-95-1381
Seifert KA, McMullen CR, Yee D, Reeleder RD, Dobinson KF (2003) Molecular differentiation and detection of ginseng-adapted isolates of the root rot fungus Cylindrocarpon destructans. Phytopathology 93:1533–1542. https://doi.org/10.1094/PHYTO.2003.93.12.1533
Sun Y, Ke J, Ma N, Chen Z, Wang C, Cui X (2004) Effects of root rot on saponin content in Pananx notoginseng. Journal of Chinese Medicinal Mater 27:79–80
Wang YH, Qiu C, Wang DW, Hu ZF, Yu BY, Zhu DN (2011) Identification of multiple constituents in the traditional Chinese medicine formula Sheng-Mai San and rat plasma after oral administration by HPLC-DAD-MS/MS. J Pharm Biomed Anal 54:1110–1127. https://doi.org/10.1016/j.jpba.2010.11.034
Watanabe M, Sumida N, Yanai K, Murakami T (2004) A novel saponin hydrolase from Neocosmospora vasinfecta var. vasinfecta. Appl Environ Microbiol 70:865–872
Winnenburg R, Urban M, Beacham A, Baldwin TK, Holland S, Lindeberg M, Hansen H, Rawlings C, Hammond-Kosack KE, Kohler J (2007) PHI-base update: additions to the pathogen host interaction database. Nucleic Acids Res 36:D572–D576. https://doi.org/10.1093/nar/gkm858
Wu Z, Hao Z, Zeng Y, Guo L, Huang L, Chen B (2015) Molecular characterization of microbial communities in the rhizosphere soils and roots of diseased and healthy Panax notoginseng. Antonie Van Leeuwenhoek 108:1059–1074. https://doi.org/10.1007/s10482-015-0560-x
Yan XL, Xiao FH, Wen GS, Li X, Duan CL (2006) Investigation and collection of wild Panax species in Yunnan province. J Yunnan Agric Univ 21:419–423
Yousef LF, Bernards MA (2006) In vitro metabolism of ginsenosides by the ginseng root pathogen Pythium irregulare. Phytochemistry 67:1740–1749. https://doi.org/10.1016/j.phytochem.2005.06.030
Zhu B, Wang S, Mi C, Yang R, Zen G, Hu X (2019) Genome sequence resource for Ilyonectria mors-panacis, causing rusty root rot of Panax notoginseng. Mol Plant Microbe Interact 32:1468–1471. https://doi.org/10.1094/MPMI-05-19-0118-A
Acknowledgements
This research is supported by the general scientific research project from Education Department of Zhejiang Province (Y201840250), general scientific research project from Zhejiang Sci-Tech University (18042289-Y), Zhejiang Natural Science Foundation of China (LQ20C010006). We are grateful to Dr. Hu for their initial experiments on this project.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
We declare that we have no conflicts of interest in the authorship or publication of this contribution.
Additional information
Communicated by Erko Stackebrandt.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zeng, G., Li, J., Ma, Y. et al. Analysis of saponins detoxification genes in Ilyonectria mors-panacis G3B inducing root rot of Panax notoginseng by RNA-Seq. Arch Microbiol 203, 5205–5213 (2021). https://doi.org/10.1007/s00203-021-02502-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-021-02502-4