Abstract
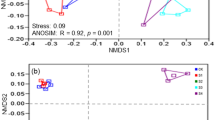
Understanding the salt tolerance of microbial communities may help to elucidate the effects of salt concentration and other environmental factors on soil biodiversity. Here, high-throughput sequencing of 16S rDNA and ITS was combined to investigate the distribution and salt tolerance of microbial communities in coastal soils and sediments near the Yinggehai saltern field of Hainan Island, China. The microbial communities in the soils and sediments of the land zone (YGHLS), the intertidal zone (YGHIS), and the inshore zone (YGHWS) were compared. PCoA of weighted and unweighted UniFrac distance revealed obvious differences in soil microbial community among different samples. ANOSIM analysis could clearly separate the three samples from each other. Three halotolerant bacteria, including Halomonas, Halobacillus and Wallemia, were found in the samples, which accounted for 0.0335 ± 0.0586%, 0.0241 ± 0.0304%, and 0.0308 ± 0.0445% of the total microbial community, respectively. The relative abundance of Trk system potassium uptake protein, Kdp operon response regulator, and Na+/H+ antiporter in the samples exceeded 0.09%, 0.06%, and 0.02%, respectively, indicating that the Trk system plays a major role in the salt tolerance of halotolerant bacteria in Yinggehai coastal soils and sediments.






Similar content being viewed by others
References
Boyadzhieva I, Tomova I, Radchenkova N, Kambourova M, Poli A, Vasileva-Tonkova E (2018) Diversity of heterotrophic halophilic bacteria isolated from coastal solar salterns, Bulgaria and their ability to synthesize bioactive molecules with biotechnological impact. Microbiology 87(4):519–528
Canini F, Geml J, D’Acqui LP, Selbmann L, Onofri S, Ventura S, Zucconi L (2020) Exchangeable cations and pH drive diversity and functionality of fungal communities in biological soil crusts from coastal sites of Victoria Land, Antarctica. Fungal Ecol. https://doi.org/10.1016/j.funeco.2020.100923
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Meth 7:335–336
Chen YG, Zhang ZH, Luo HW, Li Z, Zhang LJ, Huang H (2020) Distinct characteristics of bacterial community in the soil of Nanshazhou Island, South China Sea. Curr Microbiol 77:1292–1300
Clarke KR (1993) Non-parametric multivariate analyses of changes in community structure. Austral Ecol 18:117–143
Dini-Andreote F, Silva MCP, Triadó-Margarit X, Casamayor EO, van Elsas JD, Salles JF (2014) Dynamics of bacterial community succession in a salt marsh chronosequence: evidences for temporal niche partitioning. ISME J 8:1989–2001
Edgar RC (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461
Gunde-Cimerman N, Ramos J, Plemenitas A (2009) Halotolerant and halophilic fungi. Mycol Res 113:1231–1241
Hong YH, Huang Y, Wu S, Yang XZ, Dong YZ, Xu DY, Huang ZQ (2020) Effects of imidacloprid on the oxidative stress, detoxification and gut microbiota of Chinese mitten crab, Eriocheir sinensis. Sci Total Environ. https://doi.org/10.1016/j.scitotenv.2020.138276
Jorgensen SL, Hannisdal B, Lanzén A, Baumberger T, Flesland K, Fonseca R, Øvreås L, Steena IH, Thorseth IH, Pedersen RB, Schleper C (2012) Correlating microbial community profiles with geochemical data in highly stratified sediments from the Arctic Mid-Ocean Ridge. Proc Natl Acad Sci 109:2846–2855
Kang HZ, Gao HH, Yu WJ, Yi Y, Wang Y, Ning ML (2018) Changes in soil microbial community structure and function after afforestation depend on species and age: case study in a subtropical alluvial island. Sci Total Environ 625:1423–1432
Kraegeloh A, Amendt B, Kunte HJ (2005) Potassium transport in a halophilic member of the Bacteria domain: identification and characterization of the K+ uptake systems TrkH and TrkI from Halomonas elongata DSM 2581T. J Bacterial 187(3):1036–1043
Langille MGI, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Thurber RLV, Knight R, Beiko RG, Huttenhower C (2013) Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 31:814–821
Lozupone C, Knight R (2005) UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71:8228–8235
Lozupone C, Hamady M, Knight R (2006) UniFrac—an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics 7:371
Magoc T, Salzberg SL (2011) FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27:2957–2963
Ng ZY, Tan GYA (2018) Selective isolation and characterisation of novel members of the family Nocardiopsaceae and other actinobacteria from a marine sediment of Tioman Island. Antoni Leeuw Int J G 111(5):727–742
Osman JR, Regeard C, Badel C, Fernandes G, DuBow MS (2019) Variation of bacterial biodiversity from saline soils and estuary sediments present near the Mediterranean Sea coast of Camargue (France). Anton Leeuw Int J G 112(3):351–365
Somboonna N, Assawamakin A, Wilantho A, Tangphatsornruang S, Tongsima S (2012) Metagenomic profiles of free-living archaea, bacteria and small eukaryotes in coastal areas of Sichang island, Thailand. BMC Genomics 13:S29
Sottorff I, Wiese J, Imhoff JF (2019) High diversity and novelty of Actinobacteria isolated from the coastal zone of the geographically remote young volcanic Easter Island, Chile. Int Microbiol 22(3):377–390
Tang J, Zheng AP, Bromfield ESP, Zhu J, Li SC, Wang SQ, Deng QM, Li P (2011) 16S rRNA gene sequence analysis of halophilic and halotolerant bacteria isolated from a hypersaline pond in Sichuan, China. Ann Microbiol 61:375–381
Trigui H, Masmoudi S, Brochier-Armanet C, Barani A, Grégori G, Denis M, Dukan S, Maalej S (2011) Characterization of heterotrophic prokaryote subgroups in the Sfax coastal solar salterns by combining flow cytometry cell sorting and phylogenetic analysis. Extremophiles 15:347–358
Verma P, Raghavan RV, Jeon CO, Lee HJ, Priya PV, Dharani G, Kirubagaran R (2017) Complex bacterial communities in the deep-sea sediments of the Bay of Bengal and volcanic Barren Island in the Andaman Sea. Mar Genomics 31:33–41
White TJ, Bruns T, Lee S, Taylor JW (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols: a guide to methods and applications. Academic Press, New York, pp 315–322
Xiong W, Li R, Ren Y, Liu C, Zhao Q, Wu H, Jousset A, Shen QR (2017) Distinct roles for soil fungal and bacterial communities associated with the suppression of vanilla Fusarium wilt disease. Soil Biol Biochem 107:198–207
Yang LF, Jiang JQ, Zhao BS, Zhang B, Feng DQ, Lu WD, Wang L, Yang SS (2006) A Na+/H+ antiporter gene of the moderately halophilic bacterium Halobacillus dabanensis D-8T: cloning and molecular characterization. FEMS Microbiol Lett 255:89–95
Zalar P, de Hoog GS, Schroers HJ, Frank JM, Gunde-Cimerman N (2005) Taxonomy and phylogeny of the xerophilic genus Wallemia (Wallemiomycetes and Wallemiales, cl. et ord. nov.). Anton Leeuw Int J G 87(4):311–328
Zaura E, Keijser BJF, Huse SM, Crielaard W (2009) Defining the healthy “core microbiome” of oral microbial communities. BMC Microbiol 9:259
Zeglin LH, Wang B, Waythomas C, Rainey F, Talbot SL (2016) Organic matter quantity and source affects microbial community structure and function following volcanic eruption on Kasatochi Island, Alaska. Environ Microbiol 18:146–158
Zhang XM, Zhang Q, Liang B, Li JL (2017) Changes in the abundance and structure of bacterial communities in the greenhouse tomato cultivation system under long-term fertilization treatments. Appl Soil Ecol 121:82–89
Acknowledgements
This work was supported by Key Research and Development Projects of Hainan Province (ZDYF2019212), Natural Science Foundation of Hainan Province (318MS072) and Projects of Key Laboratory of Utilization and Conservation for Tropical Marine Bioresources, Ministry of Education (UCTMB20205).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Erko Stackebrandt.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Chen, Y., Zhang, Z., Luo, H. et al. Salt tolerance of halotolerant bacteria from coastal soils and sediments near saltern field of Hainan Island, China. Arch Microbiol 203, 5921–5930 (2021). https://doi.org/10.1007/s00203-021-02461-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-021-02461-w