Abstract
Staphylococcus aureus is responsible for numerous instances of superficial, toxin-mediated, and invasive infections. The emergence of methicillin-resistant (MRSA), as well as vancomycin-resistant (VRSA) strains of S. aureus, poses a massive threat to human health. The tenacity of S. aureus to acquire resistance against numerous antibiotics in a very short duration makes the effort towards developing new antibiotics almost futile. S. aureus owes its destructive pathogenicity to the plethora of virulent factors it produces among which a majority of them are moonlighting proteins. Moonlighting proteins are the multifunctional proteins in which a single protein, with different oligomeric conformations, perform multiple independent functions in different cell compartments. Peculiarly, proteins involved in key ancestral functions and metabolic pathways typically exhibit moonlighting functions. Pathogens mainly employ those proteins as virulent factors which exhibit high structural conservation towards their host counterparts. Consequentially, the host immune system counteracts these invading bacterial virulent factors with minimal protective action. Additionally, many moonlighting proteins also play multiple roles in various stages of pathogenicity while augmenting the virulence of the bacterium. This has necessitated elaborative studies to be conducted on moonlighting proteins of S. aureus that can serve as drug targets. This review is a small effort towards understanding the role of various moonlighting proteins in the pathogenicity of S. aureus.
Similar content being viewed by others
Introduction
In 1880, Dr Alexander Ogston had constantly noticed the presence of Gram-positive spherical “micrococci” in the pus collected from 88 human abscesses. These isolated bacteria were capable of recreating the abscesses in healthy guinea pigs and mice upon injection. This preliminary effort by Dr Ogston paved the way to the understanding of an infectious agent, now known as S. aureus, which continues to be a massive threat to human health and wellbeing (Ogston 1984). It acts as a commensal of the normal human microbiota and is asymptomatically carried by humans (20–40%), frequently found in skin flora, in the nostrils, and is a normal inhabitant of the lower female reproductive tract and anterior nares being the major sites of colonization. S. aureus has evolved as a highly infectious entity along with the emergence of numerous antibiotic-resistant strains. Over the past century, S. aureus has become a great threat to public health due to it being the major infectious agent for both nosocomial and hospital-acquired infections (Periasamy et al. 2012). S. aureus infections can be classified into superficial infections, toxin-mediated infections, and invasive infections. S. aureus causes superficial lesions ranging from milder pimples and boils to severe infection such as stys, abscesses, carbuncles and so on. Immediately after the penetration of skin barrier, S. aureus is capable of causing initial muscular and skeletal infections such as osteomyelitis and septic arthritis. These internal infections can further lead to more serious conditions of pneumonia, bacteraemia, endocarditis and septicaemia (Cramton et al. 1999; Ando et al. 2004; Anderson et al. 2012).
S. aureus is also a causative agent for numerous toxin-mediated infections (food poisoning, toxic shock syndrome and scalded skin syndrome) mainly due to its ability to produce a broad spectrum of toxins such as exfoliative toxins, superantigens and cytotoxic toxins (Ansari et al. 2014). In line with this, S. aureus is majorly correlated with skin and soft tissue infections (SSTI) both in community-mediated or invasive infections in hospitalized patients. The hospital-acquired infections of S. aureus range from ventilator-associated pneumonia to device-related infections (urinary catheters, endotracheal tubes, intravascular, prosthetic implants and arterial stents) (Anderson et al. 2012). Along with this, the emergence of methicillin-resistant (MRSA), as well as vancomycin-resistant (VRSA) strains, pose a breach in the last line of antibiotic defence (Lowy 1998; Ruffing et al. 2012). The tenacity of S. aureus to acquire resistance against numerous antibiotics in a very short duration makes the effort towards developing new antibiotics almost futile. The destructive pathogenicity of S. aureus stems from the ability to produce a plethora of virulent factors (Chhatwal 2002). Numerous studies have deciphered that most of the virulent factors of S. aureus are moonlighting proteins and they enormously potentiate the infectivity of the pathogen. This has necessitated elaborative studies on moonlighting proteins of S. aureus that can be utilized as effective drug targets.
Concept of moonlighting proteins
Numerous proteins are multifunctional mainly due to gene fusions and pleiotropic effects. But if any protein exhibits multifunctionality that cannot be ascribed to the above-mentioned processes, then they are termed as moonlighting proteins. They are the multifunctional proteins where a single protein performs multiple independent functions using different regions of the protein structure, or alternative structure (alternative structure may be attributed to post-translational modifications and/or oligomerization and/or conformational changes due to binding of different ligands). Moonlighting proteins often execute multiple functions in different cell compartments at different times (Copley 2012). A good analogy to moonlighting proteins was well explained in the review paper by Henderson et al. 2011 as being a person having two jobs, one in the day and one at night (Henderson et al. 2011). Moonlighting proteins are mainly found in plants, mammals, worms, yeast, bacteria, archaea, and viruses (Jeffery 1999).
A switch in the protein’s canonical function to its moonlighting function is mainly determined by cellular localization, cell type, oligomeric state and sometimes the cellular concentration of a ligand, substrate, cofactor or product. For instance, PutA protein of Escherichia coli which is associated with the plasma membrane acts as a proline dehydrogenase and pyrroline-5-carboxylate dehydrogenase but the same protein in the cytoplasm acts as a transcriptional repressor (Muro-Pastor et al. 1997). On the other hand, thymidine phosphorylase exhibits two distinctive functions inside and outside the cell. In the cytoplasm, the protein dephosphorylates thymidine, deoxyuridine and their analogs into their bases and 2-deoxyribose 1-phosphate. The same protein in the extracellular fluid, however, serves as a platelet-derived endothelial growth factor and also mediates chemotaxis of platelets (Furukawa et al. 1992). The human glyceraldehyde-3-phosphate dehydrogenase is a 37-kDa glycolytic enzyme which converts glyceraldehyde 3-phosphate to 1,3-bisphosphoglycerate in its tetrameric conformation. In its monomeric form, it acts as nuclear uracil–DNA glycosylase which removes uracil from DNA because of accidental use of dUTP during replication (Meyer et al. 1991). In some scenarios, moonlighting proteins exert multiple functions by utilizing different distinct binding sites. For example, the aspartate receptor of E. coli which plays a key role in chemotaxis also acts as a receptor for maltose-binding protein (MBP). But this receptor has different but overlapping binding sites for both of the ligands (Mowbray et al. 1990). Along with this, the protein may utilize its different subunits for excreting moonlighting functions. Ribonucleotide reductase (RNR) is an important enzyme that mediates the conversion of ribonucleotides to deoxyribonucleotides, the DNA precursors. This reaction is essential for DNA biosynthesis during S-phase of the cell cycle and DNA repair mechanisms (Reichard et al. 1961; Hofer et al. 2012). Numerous pieces of evidence have recently come to light which suggests that ribonucleotide reductase (RNR) exhibits moonlighting behaviors. RNR dual-subunit enzyme is composed of α- and β-subunit. RNR α-subunit serves as nucleotide-binding subunit and involves in catalytic activity and also allosterically regulates the cellular nucleotide pool for RNR conical activity (Nordlund and Reichard. 2006). The RNR-β subunit serves as a major plasminogen-binding proteins of S. aureus (Mölkänen et al. 2002).
The survival of an organism necessitates the utilization of whatever resource is available at hand. Therefore, ubiquitous and multifunctioning metabolic enzymes were the first choice to be put to work as moonlighting proteins. Majority of proteins and enzymes have a large solvent-exposed surface area. A lesser surface area is required for performing a single function that requires a single binding site. Rest of the unused surface area can be modified to create an additional binding site as long as the primary function (binding) is not compromised. This kind of evolutionary approach is beneficial as there are fewer proteins required to be synthesized and, consequently less DNA to be replicated. This significantly reduces the molecular burden on the organism and saves a great deal of energy and makes the genome more compact (Suzuki et al. 1997; Piatigorsky 1998). Franco-Serrano et al. 2018a, b, collected 698 moonlighting proteins from MultitaskProtDB-II database and analyzed them for their moonlighting and canonical functions. Interestingly, they observed that proteins involved in key ancestral functions and metabolic pathways typically exhibited moonlighting functions. Out of all studied proteins, about 25% of the proteins of the database exhibited virulence activity as a corresponding moonlighting function. Based on this peculiar observation, they hypothesized that pathogens mainly employ those proteins as virulent factors which exhibit high structural conservation towards their host counterparts. Due to this, the host immune system will elicit minimal protective action against these invading bacterial virulent factors (Franco-Serrano et al. 2018a, b).
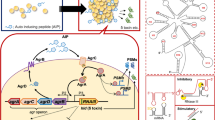
These moonlighting proteins are intracellular proteins without any canonical signal peptide or secretion motifs. But a majority of moonlighting proteins are translocated to the external cell surface for their pathogenic functions. Many mechanisms are being studied to explain this process. Cells do not take up moonlighting proteins from cells undergoing cell leakage or cell death as explained by many old theories. For example, in S. aureus, the enzymes enolase and aldolase are highly secreted to the outer surface during exponential growth and in this phase of growth, cells undergo minimum cell breakage or cell death (Ebner et al. 2015). Instead, secretion of cytoplasmic proteins is a selective and regulated process. For instance, reversible interconversion of 2‐phosphoglycerate to phosphoenolpyruvate involves the formation of a transient covalent bond between the substrate (2‐phosphoglycerate) and the active site Lys341 of the enzyme (enolase). Mutation of this amino acid residue not only hindered substrate binding but also prevented extracellular exportation of enolase (Boël et al. 2004). S. aureus possesses a plethora of moonlighting proteins which helps in pathological invasion and the subsequent establishment of the infectious cycle. The role of moonlighting proteins in the virulence of S. aureus has been depicted in the Fig. 1 and Table 1. But research on moonlighting proteins of S. aureus is at its nascent stage and the available information is relatively sparse.
Schematic diagram depicting the role of reported moonlighting proteins in the virulence of S. aureus. The figure describes the roles of moonlighting proteins in adhesion such as Autolysin, Cna, Enolase, etc. which help in the binding of host extracellular matrix proteins. Invasins such as Mnt.C, FnBP, IMPDH, etc. help in the invasion of the host system by exploiting the plasminogen-mediated fibrinolysis. GAPDH also serves as an elusive receptor for transferrin and also plays a part in NO neutralization and oxidative stress management. Autolysin also serves its prime role in the secretion of cytoplasmic proteins. TPI binds to the capsular polysaccharide of fungal pathogen C. neoformans and initiates apoptosis of the fungal pathogen. TPI also regulates the invasion with the help of Enolase. S. aureus employs some moonlighting proteins such as LipA, GAPDH, Actin, etc. in immunomodulation. Some moonlighting proteins such as Aconitase, CysK, etc. help in gene regulation
Inosine 5′-monophosphate dehydrogenase as a plasminogen receptor on the cell surface of S. aureus
Inosine 5′-monophosphate dehydrogenase (IMPDH) mainly catalyzes the nicotinamide adenine dinucleotide (NAD+)-dependent oxidation of inosine monophosphate (IMP) to xanthosine monophosphate (XMP). This is the penultimate and rate-limiting step in the de novo synthesis of guanine nucleotide. Due to this, IMPDH canonically plays an important role in nucleic acid metabolism and its subsequent regulation (Zhang et al. 1999). Apart from its metabolic contributions, IMPDH also functions as a plasminogen receptor on the cell surface of S. aureus. Plasminogen (PLG: − 90 kDa) is considered the apo-enzyme of plasmin. Plasmin is the serine protease involved in intravascular fibrinolysis and degradation of the extracellular matrix. Plasminogen is a single-chained protein composed of 3 domains—activation domain of nearly 8 kDa, five homologous disulfides bonded triple loop of 1–5 Kringle domain of nearly 65 kDa, and serine protease domain (17 kDa). The zymogen plasminogen is converted to plasmin by cleavage at the Arg561–Val562 position by tissue-type or urokinase-type plasminogen activator (tPA and uPA, respectively) (Miyashita et al. 1988). Apart from well-characterized plasminogen activators, there are many nonclassical plasminogen activators produced by the bacteria. S. aureus produces staphylokinase, a nonenzymatic chemical species which forms an equimolar stoichiometric complex with plasminogen. This association activates plasminogen to plasmin (Lijnen et al. 1991). The plasminogen activation by staphylokinase is well regulated by α2-antiplasmin l (Lottenberg et al. 1994). But these bacterially expressed plasminogen activators are unable to act on plasminogen unless bacteria bind to plasminogen. Thus, bacterium expresses surface receptors like IMPDH which first binds to plasminogen and then using plasminogen activators, it converts plasminogen to plasmin.
The active plasmin can directly hydrolyze matrix proteins such as laminin and fibronectin. It can also activate the enzymes that are involved in the complement cascade. Along with this, plasmin can activate proenzymes such as collagenases and other matrix metalloproteases that altogether disrupt the membrane integrity which results in the membrane collapse (Lottenberg et al. 1994; He et al. 1989). At this juncture, the interaction of bacterial plasminogen receptors with host plasminogen and subsequent activation to plasmin represents the correlative mechanism for invasion by the bacterium. Consecutively, IMPDH also binds to the lysine-binding site of plasminogen (1–5 kringle domain) which prevents the binding of the inhibitor α2-AP (alpha 2-antiplasmin) to activated plasmin. In this way, IMPDH not only acts as plasminogen receptor on the cell surface of S. aureus but also it protects plasmin from α2-AP-mediated inhibition (Mölkänen et al. 2002). The rapid proliferation of S. aureus in the host mainly depends on the ample quantity of guanine nucleotide pool which is regulated by IMPDH. Thus, the inhibition of IMPDH can be a successful strategy in the prevention of invasion and subsequent proliferation of S. aureus in the host. The bacterial IMPDH possess structurally distinct pocket that is absent in eukaryotic IMPDHs which can be utilized as a site for IMPDH inhibition (Hedstrom et al. 2011). For example, mycophenolic acid, an MDPH-targeted immunosuppressive drug was found to inhibit the proliferation of S. aureus (Abraham 1945). VRSA exhibits conspicuous variations in growth characteristics including high cell wall biosynthesis, generation of numerous small colony variants (SCV) and increased biofilm formation. These variations are very well correlated with upregulation of various genes involved in purine metabolism including the central enzyme IMPDH. IMPDH catalyzes
the conversion of IMP to XMP which further gets converted to guanosine monophosphate (GMP). GMP plays a critical role in energy transitions during bacterial cell wall synthesis. Further studies have shown that purines in combination with sugars also participate in the synthesis of polysaccharide components of the cell wall and the capsules which are the key events of biofilm formation. This function serves as a link between purine metabolism and pathogenesis (Macdonald and Smith 1984; Mongodin et al. 2003).
Role of glyceraldehyde-3-phosphate dehydrogenase in iron uptake, invasion and nitric oxide (NO) neutralization
The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) in S. aureus plays a prime role in glycolysis by converting glyceraldehyde 3-phosphate to 1,3-bisphosphoglycerate. S. aureus contains two homologues of GAPDH, known as GapA and GapB, with 40% overall sequence identity. GapA plays a crucial role in glycolysis while GapB is important in gluconeogenesis. Both GapA and GapB impart a significant role in the pathogenesis of S. aureus (Modun et al.1998; Goji et al. 2004; Purves et al. 2010). GAPDH exhibits various moonlighting functions in different cell compartments which have been depicted in Fig. 2. In humans, the presence of assimilable iron (free ionic iron) is highly limited because of three main reasons: first, iron is majorly found to be bound to an organic moiety (transferrin, lactoferrin, hemoproteins, etc.); second, the low solubility of iron at physiological pH and lastly, the intercellular localization of iron. This functions as a potent innate defence mechanism against bacterial infection as they require a high amount of iron for pathogenesis, approximately 0.4–4.0 μM level which is well above the physiological available iron concentration in the human body. But this defence is not failproof because the bacteria has developed high-affinity iron-scavenging mechanisms to overcome these iron limitations (Weinberg 1978; Bullen et al. 1999; Skaar and Schneewind 2004). S. aureus employs GAPDH as an elusive receptor for transferrin (Henderson et al. 2011). Pancholi and Fischetti 1992, reported that GAPDH on the cell surface of S. aureus binds to human transferrin that serves as a potential means by which bacterium can access iron in vivo (Pancholi and Fischetti 1992). This observation was further supported by Modun and Williams (1999). But, the exact mechanism by which the GAPDH helps in the acquisition of iron from the transferrin is not yet apparent. Previous studies suggest that organic phosphates (1,2-diphosphoglycerate) can mediate the release of iron from transferrin (Morgan and Brown 1977). So, 1,3-diphosphoglycerate formed by the GAPDH is capable of releasing iron bound to transferrin. Preliminary experiments suggest that 1,3-diphosphoglycerate can act as an iron scavenger and remove iron from transferrin (Modun et al.1998). But recently Taylor and Heinrichs 2002, observed that the S. aureus strains lacking the gap gene (encoding GAPDH) exhibited equal levels of transferrin binding. This observation raised a dispute upon the role of GAPDH as a transferrin receptor. Further, they also discovered a new transferrin-specific receptor (named as StbA) in S. aureus (Taylor and Heinrichs 2002). Consequently, some additional work is required to clarify the role of GAPDH as a transferrin receptor.
The canonical and moonlighting functions of GAPDH in the virulence of S. aureus are illustrated in this figure. Canonically, GAPDH has a prime role in the glycolysis and gluconeogenesis. The moonlighting function comprises of roles in NO neutralization and prevention of host-mediated oxidative stress. GAPDH is transported outside the cell with the help of Autolysin and while on the cell surface it helps in adhesion, invasion and iron acquisition
Along with this, Cole et al. 2010, demonstrated that GAPDH facilitates the adhesion and subsequent invasion of S. aureus into the corneal cells. They also observed that the S. aureus infection in corneal cells reduced upon antibody-mediated prevention of GAPDH binding to corneal epithelial cells (Cole et al. 2010). Further, GAPDH is also found to be involved in plasminogen binding (Glenting et al. 2013). Ebner et al. 2016, found that GAPDH can bind sub-fragment vitronectin, plasminogen and specific fibrinogen chains but it was unable to bind to fibronectin. They also observed that GAPDH can bind to certain host matrix proteins and enhance the adhesion but GAPDH insignificantly contributed towards host cell invasion (Ebner et al. 2016). Ebner et al. 2016, observed that GAPDH also binds to autolysin (Atl) which is one of the main moonlighting proteins on the cell surface of S. aureus that is mainly involved in the host cell internalization of S. aureus (Ebner et al. 2016).
Upon activation, macrophage can inhibit pathogen growth by producing numerous effector molecules such as Nitric oxide (NO). NO is the part of toxic molecule defence system which also regulates the growth, functional activity and senescence of many immune cells (T lymphocytes, macrophages, mast cells, neutrophils and Natural killer cells) (Bogdan 2001). Counterintuitively, S. aureus contains nitric oxide synthases whose functions is to produce NO during infection and potentiates the infectivity of S. aureus (Chartier and Couture 2007). Recently, the utilization of GAPDH by S. aureus for circumvention of NO toxicity and strategies to utilize NO–GAPDH complex as a counterattack measure to forestall the macrophage-mediated bacterial cell damage and destruction was extensively studied (Benhar and Stamler 2005). The bacterial production of GAPDHcys–NO complex (GAPDH is nitrosylated at its active site cysteine) can efficiently initiate pleiotropic changes in infected cells such as apoptosis, regulation of gene expression (via transcriptional and post-translational mechanisms) and regulation of heme synthesis via Fe++ metabolism. Along with this, GAPDHcys–NO also can actively transfer its nitroso group to other acceptor proteins (Sirover 2011, 2012, 2014; Tristan et al. 2011). In both prokaryotic and eukaryotic cells, the reduced glutathione (GSH) serves as the most abundant cellular redox buffer (1–10 mM) (Brandes et al. 2009; Van Laer et al. 2013). Apart from these, S. aureus utilizes GAPDH, one of the abundant cytosolic proteins (5–20%), for S-thiolation and subsequent circumvention of oxidative stress. The catalytic site of the GAPDH has conserved cysteine at 149th position (Cys149). Cys149 binds to thiohemiacetal intermediate of GAPDH substrate (Corbier et al. 1994). This binding affinity makes Cys149 susceptible to S-thiolation (Imber et al. 2018). The S-thiolation and subsequent inactivation of GAPDH were observed when GAPDH was incubated with glutathione disulfide (GSSG) and H2O2 along with GSH in vitro (Zaffagnini et al. 2007; Barinova et al. 2017). The S-thiolation of GAPDH results in termination of glycolysis and activation of hexose monophosphate shunt pathway (pentose–phosphate pathway). This pathway results in the production of reducing equivalent of NAPDH which serves as the main adaptation under oxidative stress (Ralser et al. 2007; Shenton and Grant 2003). Recently, GAPDH was also associated with the neutralization of sodium hypochlorite (NaOCl)-induced oxidative stress in S. aureus (Tsuchiya et al. 2018). Weber et al. 2004, showed a significant modification in the active site of GAPDHcys in S. aureus due to oxidative stress. In this study, the GAPDH was exposed to 100-μM H2O2 for 5 min. The exposure resulted in reversible alteration in the isoelectric point (pI) of GAPDH (from alkaline to acidic). The modified GAPDH was isolated from exposed cells and studied which indicated the sulfonation in the active site cysteine. This study is important for several reasons, I). The effect is rapid in comparison to catalase and Fenton reaction. II). There was a sharp change in the protein structure upon exposure and more amount of GAPDH converted to GAPDHcys–NO. III). The effect is reversible (Weber et al. 2004). Many studies observed that within 30min of exposure to H2O2, the bacteria revived and efficient proliferation was observed. In this time duration, ample amount of GAPDHcys–NO formation was detected and its subsequent conversion to the normal native form (upon the termination of H2O2 exposure) was observed (Benhar and Stamler 2005; Hara et al. 2005). Purves et al. 2010 infected the larvae of the greater wax moth Galleria mellonella with S. aureus and they observed that there was a significant increase in the survival of the infected larvae as a result of the loss of either of the Gap proteins individually and loss of both the GapA and GapB resulted in severe attenuation of bacteria along with complete survival of the infected larvae. Based on this study, they concluded that both GapA and GapB play an essential and closely associated role in the pathogenesis of S. aureus (Purves et al. 2010). On the contrary, the crucial data on the role of individual Gap proteins in the pathogenesis of S. aureus are sparse. So, an elaborative study is required for a clear understanding of the role of individual Gap proteins in virulence of S. aureus.
Enolase—a prime invasive protein of S. aureus
Enolase is the glycolytic metalloenzyme that catalyzes the reversible interconversion of 2-phosphoglycerate (2-PG) and phosphoenolpyruvate (PEP) and is involved in energy metabolism (Wold 1971). Wu et al. (2015) showed that S. aureus has both octameric and dimeric form of enolase. The octameric form being catalytically active resides in the cytoplasm and dimeric form being structurally stable form are located on the membrane. Further, the dimeric form was predicted to be involved in the pathogenicity of S. aureus (Wu et al. 2015) Lopes et al. (1985) deciphered the role of enolase as a laminin receptor on the cell surface of S. aureus. They observed that only the pathogenic S. aureus utilizes enolase as laminin receptor; whereas, the non-invasive S. epidermidis does not employ enolase for laminin binding nor does it show any laminin-binding capacity (Lopes et al. 1985). Mölkänen et al. 2002 observed that in comparison to IMPDH and ribonucleotide reductase, enolase plays a significant role in staphylokinase-induced plasminogen activation and also prevents the α2-AP-mediated plasmin inhibition. Besides, enolase also serves as a receptor for many extracellular matrix (ECM) components. Enolase mainly binds to laminin and collagen I, but not to fibronectin and collagen IV (Antikainen et al. 2007a). Foulston et al. 2014, hypothesized that S. aureus recycles the metabolic enzymes mainly enolase, to an extracellular matrix during the biofilm synthesis. They explained that bacterial cells mainly release enolase from cytoplasm to the exterior of cells in stationary phase. Further, enolase associates with the cell surface as the pH declines during biofilm formation (Foulston et al. 2014). Studies conducted by Hajighahramani et al. 2017 have shown that the presence of multiple virulent factors makes the development of an effective vaccine against S. aureus almost futile. Thus, as an alternative approach, more than one virulent factor should be targeted for the development of a potent vaccine accompanied by appropriate adjuvant (Adamczyk-Poplawska et al. 2011). For the development of an efficient multi-epitope subunit vaccine, they evaluated all the virulent factors of S. aureus by a bioinformatic tool. Out of all the virulent factors, three main antigenic determinants [Alpha-enolase (Eno1), clumping factor A (ClfA), and iron surface determinant B (IsdB)] were found to be crucial towards induction of host immune response against S. aureus and subsequent elimination of pathogenesis (Hajighahramani et al. 2017).
Moonlighting function of triose phosphate isomerase
Triose Phosphate Isomerase (TPI) is a dimeric glycolytic enzyme which catalyzes the reversible interconversion of the dihydroxyacetone phosphate (DHAP) and D-glyceraldehyde-3-phosphate (GAP) (Davenport et al. 1991). Apart from its metabolic function, TPI also acts as a membrane adhesin. TPI was found to be interacting with host proteins such as thrombin, fibronectin, fibrinogen, and plasminogen (Mölkänen et al. 2002). The lysine residue of TPI interacts with plasminogen and the interaction can be prevented by lysine analog (ε-aminocaproic acid) (Furuya and Ikeda 2011). Interestingly, the genes for enolase, GAPDH and TRP are found on the same operon. So, a significant correlation among these proteins can be expected during invasion, colonization, and dissemination of the pathogen (Furuya and Ikeda 2009). TPI and enolase bind to plasminogen with the same affinity. Enolase induces the conversion of plasminogen to plasmin; whereas, TPI decreases the conversion of plasminogen to plasmin which indicates that TPI inhibits the fibrinolysis. Thus, TPI along with enolase regulates the fibrinolysis and fibrinogenesis thereby enhancing the invasive capability of S. aureus (Furuya and Ikeda 2011). Ikeda et al. 2007, reported that S. aureus also utilizes TPI to bind to the capsular polysaccharide of fungal pathogen C. neoformans and initiates apoptosis of the fungal pathogen (Ikeda et al. 2007).
Role of autolysins in S. aureus invasion, biofilm formation and extracellular secretion of cytoplasmic proteins
Autolysin (Atl) is a peptidoglycan hydrolase (PGHs) mainly involved in the degradation of the bacterial cell wall and also the separation of daughter cells during cell division. In S. aureus, atl gene produces an inactive precursor protein (137 kDa) containing N-acetylmuramyl-L-alanine amidase (AM) and endo-13-N-acetylglucosaminidase (GM) and endopeptidase domains. (Houston et al. 2011) This precursor protein is proteolytically cleaved by trypsin to generate distinctive PGHs, 51-kDa GM and 62-kDa AM. Apart from its classical function, Atl also binds to host proteins found on extracellular matrix and body fluids, such as fibronectin (Fn), fibrinogen (Fg), vitronectin (Vn) and heparin (He) (Heilmann et al. 2003, 2005) through which it plays an important role in the invasion of the bacterium. Previous studies observed that binding of bacterial Atl to host heparin enhances the integrity of biofilm in many S. aureus strains. Heparin is a heterogeneous glycosaminoglycan and anticoagulant that helps bacteria to adhere to polystyrene (medical devices like catheter) in a dose-dependent manner. Studies also observed that S. aureus is better able to adhere to one another in the presence of heparin (Shanks et al. 2005). In conclusion, Atl is involved in the attachment of bacteria on polystyrene surface, lysis-mediated biofilm formation and also recently shown to be involved in secretion of cytoplasmic proteins (Bose et al. 2012). S. aureus utilizes the binding property of AM and GM with the human plasma as well as extracellular proteins in implant-associated infections.
Numerous cytoplasmic proteins relocate to the cell membrane to perform moonlighting functions. Very little is known about the way these proteins travel to the cell membrane because cytoplasmic moonlighting proteins lack signal sequences or other peptide motifs for targeting themselves to the membrane. At this juncture, Pasztor et al 2010, extensively studied the role of autolysin in the secretion of cytoplasmic proteins. They observed that autolysin canonically involves in septum formation during cell division. At this stage, Atl binds to newly forming cell wall and resolves the interlinked murein layers of newly dividing cells. This implies that the septum region of dividing cells may serve as a leaky site for cytoplasmic protein due to autolysin activity (Yamada et al. 1996; Schlag et al. 2010). They also generated an atl mutant and observed the significant reduction in the secretion of cytoplasmic proteins (Pasztor et al. 2010). S. aureus primarily binds to a solid surface and initiates the biofilm synthesis. Further, the successful growth of bacteria in the biofilm is determined by the bacterial adhesion and formation of an extracellular matrix which often comprises of released DNA from a subpopulation of biofilm. Autolysin mainly determines the release of DNA from subpopulation into the matrix. When biofilm formation is undesirable then S. aureus executes V8 protease-mediated degradation of Atl by which it blocks biofilm formation (Prasad et al. 2004).
Moonlighting functions of fibrinogen and fibronectin-binding proteins A and B
Fibronectin-binding protein A and B (FnBPA and FnBPB) are multifunctional proteins mainly found on the cell membrane and cytoplasm of the bacteria. The N-terminal domain of FnBPA binds to fibrinogen and helps in the invasion. Whereas, the C-terminal of FnBPA has unstructured fibronectin-binding repeats which act as the stalk for binding fibronectin (Loughman et al. 2008). N-terminal of both FnBPA and FnBPB has A-domain (structurally similar between two) which helps in binding to elastin, a key protein of the extracellular matrix mainly found in connective tissue (Deivanayagam et al. 2002). Invasion of mammalian cells by S. aureus is facilitated by a fibronectin bridge between α5β1 integrin of the host cell and FnBPs of bacterium (Sinha et al. 1999). Interaction of host α5β1 integrin and bacterial FnBPs on cell surface induces integrin clustering. This process further initiates signaling via Focal adhesion kinase (FAK) and steroid receptor coactivator (Src) kinases in association with activation of tyrosine kinase and reorganization of the actin cytoskeleton. The above-mentioned signaling cascade promotes bacterial adhesion, invasion, survival, proliferation and motility (Agerer et al. 2003). S. aureus utilizes FnBPA as a regulatory element which regulates the invasion of S. aureus into the host endothelial cells. It has been observed that bacteria do not initiate the invasion process until the bacterial toxins get biologically active. In that case, S. aureus utilizes the FnBPA to initiate actin- and Rab5-determined cytoskeletal reorganization to expel the already internalized bacteria to move to outer plasma membrane surface while preventing further uptake of bacterium (Schröder et al. 2006).
Platelets play a significant role in innate immunity. Platelets express many innate immune receptors (Toll-like receptors (TLRs), thrombin receptors and complement receptors) on its surface. Thus, platelets are involved in the identification of molecular features of microbes and subsequent recruitment of immune cells. Along with this, platelets also produce numerous anti-microbial molecules (defensins, thrombocidins and kinocidins) (Semple et al. 2011). Due to this, S. aureus mainly targets platelets and causes platelet aggregation. In S. aureus, FnBPA induces the platelet aggregation by binding to GPIIb/IIIa on the resting platelet (Fitzgerald et al. 2006).
One of the peculiar features of both FnBPA and FnBPB is that they can engage in homophilic interactions by which they form homodimers of FnBPA or homodimers of FnBPB. Due to this, they greatly contribute towards S. aureus biofilm formation as two adjacent FnBP molecules of neighboring cells can bind together (Geoghegan et al. 2013). Recently, healthcare-associated MRSA strain BH1CC and community-associated MRSA strain LAC were observed to be dependent on FnBPs for biofilm formation (Pozzi et al. 2012; Agarwal et al. 2013). Apart from these activities, FnBPs were also found to be involved in plasminogen binding as well as the conversion of plasminogen to plasmin using endogenous staphylokinase. Two evolutionary conserved lysine residues on the surface of FnBPs mediate the interaction of FnBPs to kringle 4 of the plasminogen (Pietrocola et al. 2016). FnBPs activates plasmin for two important reasons, (1) Plasmin helps in invasion as already discussed in previous examples. (2) S. aureus is well known for targeting the complement system (part of innate immune responses) because activation of the complement system by host results in microbial opsonization, leukocyte recruitment, and cell lysis. The activated C3 is (anaphylatoxin C3a and opsonin C3b) the key component of the complement system as C3a attracts and activates granulocytes, C3b leads bacteria to phagocytosis and also creates pores on the bacterial cell surface (Walport 2001). Plasmin can cleave native C3 and generates unstable anaphylatoxin C3a and C3b which is subsequently inactivated to form iC3b and C3c and later degraded. Along with this, FnBPs also inhibits the binding of Factor B to C3b by which it can successfully block the C3 and C5 convertases. All these mechanisms help S. aureus to maximize complement inhibition at the C3 level itself (Jongerius 2007; Haspel et al. 2008).
Cytoskeletal proteins as a moonlighting protein in S. aureus
S. aureus has long been considered as a non-intracellular pathogen. But recent studies revealed that the bacterium remains active in phagocytotic bodies. Along with this, it can survive intracellularly in epithelial and endothelial cells (Bayles et al. 1998). Recent studies have shown that actin polymerisation has been utilized by S. aureus as the driving force for intracellular movements, dissemination within infected tissue and cell-to-cell spread. Jung et al. 2001, observed the direct correlation between actin density in the host and invasiveness of S. aureus to oral epithelial cells (Jung et al. 2001). It has been observed that inside the infected cells, the continuous actin filament polymerization and depolymerization drive the propulsion of bacteria (Lasa et al. 1997). Staphylococcal protein A (SPA) performs multiple functions in the pathogenesis of S. aureus. For instance, it binds to the Fc region of the immunoglobulin and involves in immunomodulation. Along with this, many studies showed that the actin also binds to SPA in renal epithelial cells and is indirectly correlated in invasive behavior of pathogens (Meyer et al. 1991; Mintz and Fives-Taylor 1994).
Collagen (Cn)-binding protein as an immune evasion factor and adhesion factor
Collagen (Cn) is the major glycoprotein found in connective tissue. The staphylococcal collagen (Cn)-binding protein (Cna) binds to type I collagen (CNI) which is one of the major organic components of the human body. Collagen (Cn)-binding protein (Cna) is the prime prototype of MSCRAMMs (acronym for "microbial surface components recognizing adhesive matrix molecules”—bacterial proteins which mediate the initial adhesion on the host cell) and it acts as both immune evasion factor and adhesion factor due to which it significantly contributes to staphylococcal pathogenicity. In septic arthritis, Cna adhesion to collagen was directly correlated with disease pathogenesis (Nilsson et al. 1998). Besides this, in some studies, the Cna is involved in the prevention of the classical pathway of the complement system by binding complement protein C1q (Kang et al. 2013). Patti et al. 1994, infected one set of mice with Cna-deficient S. aureus (group A) and another set of mice with Cna-expressing S. aureus (group B). A significant reduction in the incidence of arthritis was observed in group A in comparison to group B (Patti et al. 1994). Furthermore, Nilsson et al. 1998, observed that the vaccination of mice by a recombinant fragment of Cna protected the mice from septic death even after being injected with a lethal dose of S. aureus (Nilsson et al. 1998). This study highlighted the involvement of Cna in the S. aureus-mediated arthritis. Some studies also showed that Cna-deficient S. aureus strains have the lower capability in inducing keratitis (Rhem et al. 2000), endocarditis (Hienz et al. 1996) and osteomyelitis (Elasri et al. 2002). Valotteau et al 2017, observed that Cna can also have multivalent or cooperative interactions host laminin.
Lipoic acid synthetase: the inhibitor of macrophage
Lipoic acid is a cofactor for numerous key enzyme complexes of oxidative and one-carbon metabolism. The de novo synthesis of lipoic acid is mainly performed by lipoic acid synthetase (LipA) (Zhang 2015). LipA has recently been studied as a moonlighting protein in the S. aureus. The prime moonlighting function of LipA is the suppression of macrophage activation in the host. A potent phagocytic leukocyte, i.e., macrophage is the key component of innate immunity. Macrophage can successfully destruct the invading pathogens by phagocytosis and oxidative-mediated cell lysis. So, efficient suppression of macrophage activation is the key process which determines the survival and pathogenicity of S. aureus in the host (Flannagan 2009). In S. aureus, LipA adds the lipoic acid group to pyruvate dehydrogenase (PDH), thus forming protein lipoyl-E2-PDH. Grayczyk et al. 2017, demonstrated that this lipoyl-E2-PDH complex prevents the lipopeptide-mediated activation of Toll-like receptor 1/2 (TLR1/2) on the macrophage. It has also been observed that in the murine systemic infection, LipA-mediated suppression of macrophages resulted in uncontrolled S. aureus infection. (Grayczyk et al. 2017). This peculiar observation highlights the corrective function of LipA in bacterial metabolism and immune evasion.
dUTPases—the de‐repressor protein of the pathogenicity islands in S. aureus
The dUTPases (dUTP pyrophosphatase; Dut; EC 3.6.1.23) are the ubiquitous enzymes that play an important role in the prevention of misincorporation of the uracil into DNA and is also involved in the regulation of cellular dUTP levels in the bacteria (Vértessy and Tóth 2009). Apart from its fundamental functions, the staphylococcal bacteriophage encoded Duts is also involved in de-repressor proteins of the pathogenicity islands of S. aureus (SaPIbov1 and SaPIbov5) (Vértessy and Tóth 2009; Kouzminova and Kouzminov 2004). Staphylococcal pathogenicity islands (SaPIs) are mobile genetic elements which carry numerous virulence factor genes such as toxic shock syndrome toxin TSST gene. In the pathogenicity islands, the phage-inducible chromosomal islands (composed of highly evolved molecular virulent genes) passively reside in the host (bacterial) chromosome. These islands are kept under the control of Stl, a global SaPI-encoded repressor protein (Penadés and Christie 2015). Upon induction by resident prophage or infection by a helper phage, the SaPIs excise itself, undergo autonomous replication and are packed into the phage particles (composed of virion proteins of phage). To activate the repressor (SaPI), certain proteins act as anti-repressor and binds to SaPI-encoded repressor Stl (Tormo-Más et al. 2010, 2013). Dut protein in its trimeric and dimeric conformation acts as anti-repressor proteins for SaPIs (subsets of SaPIs such as SaPIbov1, SaPIbov5 or SaPIov1). Dut derepresses the SaPIbov1 by direct binding to Stl thus preventing Stl from binding to DNA (Tormo-Más et al. 2010; Hill and Dokland 2016). Duts mainly employ the hydrolysis of nucleotides as a switch in SaPI derepression. The hydrolysis of dUMP nucleotide results in turning ‘off’ of this SaPI derepression. On the other hand, triphosphate nucleotide dUTP is responsible for the ‘on’ state of derepression (Tormo-Más et al. 2010, 2013; Carpena et al. 2016).
Manganese transport protein protects S. aureus from host-exerted oxidative stress
Manganese transport protein (MntC) is primarily a metal ion transmembrane transporter in bacteria. The prime colonizing site of S. aureus is nasopharynx. Such mucosal sites are rich in several metal ions including Manganese. So, S. aureus possesses a strong metal ion transmembrane transporter for Manganese. Apart from using manganese for vital cellular process, S. aureus also exploits manganese uptake mechanism to protect itself from the oxidative burst in the nasopharyngeal site. (Gupta et al. 2013). Handke et al. showed the role of MntC in development of resistance to host-mediated oxidative stress by competing with host calprotectin for free manganese (Handke et al. 2013‚ 2018). As a consequence, MntC pulls up all free Manganese from the host and provides manganese for superoxide dismutase (SOD) (SodA and SodM) which is directly involved in the detoxification of superoxide radicals. Coady et al. 2015 have shown that MntC mutant strain (S. aureus USA300) was more susceptible to oxidative burst by neutrophils and showed milder growth in comparison to its wild-type counterpart (Coady et al. 2015). The putative role of MntC as an adhesin to components of the extracellular matrix can be elucidated due to a significant presence of lysine residues in the C-terminal. In addition to the transporter role, MntC also binds plasminogen and is involved in the invasion and subsequent pathogenesis (Salazar et al. 2014).
Trigger enzymes of S. aureus and their role in virulence
In the battle of existence, the bacteria always effectively examines its surrounding environment and properly responds to even the subtle changes caused by the host or other bacteria. As a consequence, in bacteria, the virulence is intimately linked to the metabolism. In the course of evolution, bacteria have developed numerous intricate gene regulatory network to establish an effective correlation between metabolism and pathogenicity. Due to the extreme instability of bacterial mRNAs, the transcription initiation serves as a prime target to effectively regulate the gene expression. The expression of virulent genes is prerequisite for bacterial growth as they involve in the acquisition of nutrients. So, the expression of virulent genes is predefined by the availability of nutrients (Commichau et al. 2015). The substrate-specific enzymes are the best available source for the acquisition of numerous information on the availability of nutrients in the surroundings. A few metabolic enzymes possess DNA/RNA-binding domains and are involved in the regulation of gene expression in response to substrate availability. Such enzymes are termed as trigger enzymes. These enzymes along with their canonical functions also have secondary gene regulatory function (moonlighting function). This function is governed by the substrate availability and they transduce the signal derived by the metabolism to control the gene expression, by which they serve as a potential linker to correlate metabolism and virulence of bacteria (Greenberg 2000; Commichau et al. 2008; Commichau et al. 2015). DNA is more stable than RNA but RNA is more reactive and can attain various structural conformations. This ability allows RNA to interact with many molecules such as proteins, metabolites and other RNA (Ellington and Szostak 1992; Stülke 2002). S. aureus processes one of the unique RNA-binding trigger enzymes, i.e., aconitase (Leipuviene and Theil 2007; Volz 2008). Aconitase is a key player of the citric acid cycle or tricarboxylic acid (TCA). It is involved in the isomerization of citrate into isocitrate (Artymiuk and Green 2006). Aconitase is one of the abundant metabolic enzymes and mainly depends on the iron–sulfur cluster [solvent-exposed (Fe4–S4)] for its enzymatic activity (Beinert et al. 1996). These iron–sulfur clusters are very sensitive to intracellular iron concentrations and disassemble under iron deficiencies (Rouault and Klausner 1996). As explained in the previous part of this review, iron is usually considered as a growth-limiting nutrient for S. aureus. In the iron-deficient condition, the aconitase turns inactive due to the disassembly of the iron–sulfur cluster which in turn renders TCA cycle inoperable. The inactive aconitase predominantly serves as the RNA-binding trigger protein and binds to iron-responsive elements (IREs) in the mRNAs of genes involved in intracellular iron homoeostasis (Fig. 3) (Kaptain et al. 1991; Somerville et al. 1999). Somerville et al. 2002 genetically inactivated aconitase gene in S. aureus and observed that in the absence of aconitase as well as TCA cycle altogether, there was a significant decline in the production of several virulence factors [such as α and β toxins and glycerol ester hydrolase (lipase)] followed by premature entry into the stationary phase (Somerville et al. 2002). Along with this, Chatterjee et al. 2005 observed that aconitase inactivation in S. aureus not only reduced post-exponential cell density due to metabolic impairment but also drastically impaired the production of accessory gene regulator (agr)-dependent virulence factors (such as agr quorum-sensing system) (Chatterjee et al. 2005). Choby et al. 2016 carried out the disruption of Fe–S cluster using a small molecule inhibitor and studied its implication on the pathogenicity of S. aureus. There was a significant decrease in the activity of the Fe–S cluster-dependent enzyme aconitase and subsequent virulence gene expression (Choby et al, 2016). Walden et al. 2006 carried out the structural analysis of IRE-bound aconitase and found that in the presence of iron, the enzyme attains the stable closed conformation with the iron-sulfur cluster as a ligand. In the absence of iron, the enzyme iron–sulfur cluster dissociates from the apo-protein and attains open conformation. The unstable open conformation exposes the IRE-binding domains of the aconitase followed by binding of aconitase to IRE (Walden et al. 2006). Elaborative studies on aconitase led to the discovery of more such trigger enzymes in S. aureus. The correlation between sulfur metabolism and bacterial pathogenesis has been studied in recent years. Cysteine synthesis occurs in S. aureus by two pathways: reverse transsulfuration pathway utilizing the homocysteine as a backbone material (Hullo et al. 2007) and thiolation pathway—initially, O-acetyl-l-serine (OAS) produced from acetyl-CoA and serine by CysE, the serine acetyltransferase; finally, OAS-thiol-lyase, CysK, condenses the sulfide and OAS to form cysteine (van der Ploeg et al. 2001). Recently, numerous studies have shown the moonlighting functions of many enzymes involved in cysteine synthesis. For instance, in the absence of exogenous cysteine, CysE and CysK together forms a bienzymatic complex and is involved in cysteine synthesis (Zhao et al. 2006). But in the presence of exogenous cysteine, the bienzymatic complex formation is prevented and the free cysK then forms a complex with transcription factor CymR (Tanous et al. 2008). CymR is a dimeric transcription factor which binds to DNA as a dimer or tetramer using a helix–turn–helix motif. It has been observed that the affinity of CymR/DNA interaction increases up to sevenfold in the presence of CysK (Tanous et al. 2008). The interaction allows the expression of many genes that are involved in cysteine formation and sulfur utilization (Even et al. 2006; Hullo et al. 2010).
Aconitase is a key player of the citric acid cycle or tricarboxylic acid (TCA). It is involved in the isomerization of citrate into isocitrate with the help of the iron–sulfur cluster (solvent-exposed [Fe4–S4]). These iron-sulfur clusters are very sensitive to intracellular iron concentrations and disassemble under iron deficiencies which in turn render TCA cycle inoperable. The inactive aconitase predominantly serves as the RNA-binding trigger protein and binds to iron-responsive elements (IREs) in the mRNAs of genes involved in intracellular iron homoeostasis and is also involved in the regulation of virulence gene expression
Regulation of expression of virulence factors including moonlighting proteins
S. aureus modulates its virulence depending on the host immunity and nutrient quality. So, genes encoding virulence factors are highly regulated and tightly synchronized with the biological cycle and quorum-sensing mechanisms of S. aureus. For example, the genes which encode for adhesion proteins and defence against the immune system (protein A, coagulase, fibronectin-binding proteins) of the host are upregulated at the initiation of the infection cycle followed by the production of degradative enzymes such as haemolysins, cytotoxins and proteases. The sophisticated regulatory mechanism is exerted by the accessory gene regulator (agr) two-component system in the S. aureus and S. aureus accessory element (sae) operons (Novick and Geisinger 2008). Agr mainly regulates the expression of adhesion protein and toxin-related genes (Novick and Geisinger 2008). The agr regulatory unit is composed of two divergent transcription units RNAII and RNAIII. The RNAII system regulates the initial gene expression (adhesion and invasion genes) and RNAIII stimulates the expression of extracellular toxins and enzymes (Janzon and Arvidson 1990). S. aureus has a circular chromosome of 2.8–2.9Mbp in size with 33% of G + C content (Archer and Crossley 1997). In the entire S. aureus genome, 75% is the core genome and the rest 25% consists of mobile genetic elements which possess the ability of horizontal transfer between strains. (Lindsay and Holden 2004). The mobile genetic elements like staphylococcal pathogenicity islands have genes for superantigen toxins (SaPIs) and one of the archetypes of this SaPIs family is SaPI1 that codes for toxic shock syndrome toxin TSST (tst) (Lindsay et al. 1998). SapI3 encodes enterotoxin B. In addition to SaPIs, S. aureus also possesses other families of the genomic island such as νSa family. They carry enterotoxin genes and toxic shock syndrome toxin genes (Gill et al. 2005). Also, some of the νSa family genomic islands code for leukocidin (LukDE) (Baba et al. 2002). Along with SaPIs, prophages also play an important role in the evolution and pathogenicity of S. aureus due to horizontal transfer of genetic information. Mainly three prophages have been identified in different strains of S. aureus based on the size of their genome. Most of these prophages carry virulence determinants, for example, staphylokinase, exfoliative toxin, enterotoxins A, G, K, and Panton–Valentine Leukocidin (Kuroda et al. 2001; Lindsay and Holden 2004; Diep et al. 2006). In addition to these, S. aureus also has plasmids which play a crucial role in the development of resistance to antibiotics or heavy metals, survival and adaptation in harsh environmental conditions, expression of virulence factors and survival in nutritional depletion (Wegrzyn 2005). Based on the size, the plasmids of S. aureus have been divided into three classes. Class I plasmids (1–5 kb) have a high copy number and carry antibiotic-resistance determinant. The class II plasmids have an intermediate size and copy number and they code for β-lactamase of which majority of them confer resistance to inorganic ions. The class III plasmids are large plasmids (40–60 kb) which have multiple resistance determinants (resistance to trimethoprim and gentamycin) (Novick 1989). These plasmids are means of successful transfer of antibiotic resistance by a conjugative horizontal transfer mechanism (Hartleib et al. 2000; Gill et al. 2005; Diep et al. 2006). Purine anabolic pathway is mainly regulated by purine biosynthesis repressor (PurR). Along with this PurR is also involved in the modulation of S. aureus virulence gene expression. It was observed that down-regulation of purR gene results in upregulation of genes encoding FnBPs. Goncheva et al. 2019 observed that PurR mutants exhibited hypervirulence and hypervirulence was correlated with hyper-clumping phenotype in serum (Goncheva et al. 2019).
Conclusion
S. aureus is the causative agent for multiple infections. The infectivity is mainly due to various potent virulent factors. Moonlighting proteins significantly contribute to potentiating the infectivity of S. aureus by function as virulent factors. S. aureus employs these moonlighting proteins in various steps of the infection cycle, mainly in some of the major infection steps like invasion (IMPDH, GAPDH, enolase, etc.), immunomodulation (FnBP, LipA, Cna, etc.), biofilm formation (Autolysin, enolase, etc.). One of the peculiar features of these moonlighting proteins is the important roles they play in the many steps of the infection cycle. For example, GAPDH serves as an invasin which aids the invasion process of bacteria and also helps in iron uptake as well as neutralization of the immune system induced NO toxicity. Recently, an ample amount of data have been generated regarding the functionality of various moonlighting proteins in the pathogenicity of S. aureus and the list of new moonlighting proteins is growing significantly with respect to time. But the data are more scattered and a significant correlation has not been properly established. A systematic analysis of the role of all the moonlighting proteins in the infection cycle and their introspective correlation can yield a novel approach by which a key drug element can be identified or it may aid in the development of a potent vaccine against S. aureus. Various efforts have been taken to scrutinize the data on moonlighting proteins such as the development of databases on moonlighting proteins, for example, MoonProt (https://www.moonlightingproteins.org/) (Chen et al. 2018 m). But developments on scrutinization of data available on moonlighting proteins of S. aureus are at its nascent stage and the available information is relatively sparse. Current knowledge on moonlighting proteins have cleared the fact that the age-old method of targeting a single virulence factor for vaccine development will not be fruitful as S. aureus recruits numerous multifunctional proteins in exerting its pathogenicity. So, consideration of more than two or three virulent determinates would be more promising for the development of potent vaccines. Along with this, immunizations with the plasmid containing either partial or complete protein-encoding gene followed by a boost with recombined protein may serve as a prime strategy in the prevention of S. aureus infections.
References
Abraham E (1945) The effect of mycophenolic acid on the growth of Staphylococcus aureus in heart broth. Biochem J 39:398. https://doi.org/10.1042/bj0390398
Adamczyk-Poplawska M, Markowicz S, Jagusztyn-Krynicka EK (2011) Proteomics for development of vaccine. J Proteom 74:2596–2616. https://doi.org/10.1016/j.jprot.2011.01.019
Agarwal V, Kuchipudi A, Fulde M, Riesbeck K, Bergmann S, Blom AM (2013) Streptococcus pneumoniae endopeptidase O (PepO) is a multifunctional plasminogen-and fibronectin-binding protein, facilitating evasion of innate immunity and invasion of host cells. J Biol Chem 288:6849–6863. https://doi.org/10.1074/jbc.M112.405530
Agerer F, Michel A, Ohlsen K, Hauck CR (2003) Integrin-mediated invasion of Staphylococcus aureus into human cells requires Src family protein-tyrosine kinases. J Biol Chem 278:42524–42531. https://doi.org/10.1074/jbc.M302096200
Anderson AS, Scully IL, Timofeyeva Y, Murphy E, McNeil LK, Mininni T, Nuñez L, Carriere M, Singer C, Dilts DA (2012) Staphylococcus aureus manganese transport protein C is a highly conserved cell surface protein that elicits protective immunity against S. aureus and Staphylococcus epidermidis. J Infect Dis 205:1688–1696. https://doi.org/10.1093/infdis/jis272
Ando E, Monden K, Mitsuhata R, Kariyama R, Kumon H (2004) Biofilm formation among methicillin-resistant Staphylococcus aureus isolates from patients with urinary tract infection. Acta Med Okayama 58:207–214. https://doi.org/10.18926/AMO/32090
Ansari S, Nepal HP, Gautam R, Rayamajhi N, Shrestha S, Upadhyay G, Acharya A, Chapagain ML (2014) Threat of drug resistant Staphylococcus aureus to health in Nepal. BMC Infect Dis 14:157. https://doi.org/10.1186/1471-2334-14-157
Antikainen J, Kuparinen V, Lähteenmäki K, Korhonen TK (2007) Enolases from Gram-positive bacterial pathogens and commensal lactobacilli share functional similarity in virulence-associated traits. FEMS Immunol Med Microbiol 51:526–534. https://doi.org/10.1111/j.1574-695X.2007.00330.x
Archer GL, Crossley KB (1997) The staphylococci in human disease. Churchill Livingstone, New York (NY). https://lib.ugent.be/catalog/rug01:00042699
Artymiuk PJ, Green J (2006) The double life of aconitase. Structure 14:2–4. https://doi.org/10.1016/j.str.2005.12.001
Baba T, Takeuchi F, Kuroda M, Yuzawa H, Aoki K-i, Oguchi A, Nagai Y, Iwama N, Asano K, Naimi T (2002) Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 359:1819–1827. https://doi.org/10.1016/s0140-6736(02)08713-5
Barinova KV, Serebryakova MV, Muronetz VI, Schmalhausen EV (2017) S-glutathionylation of glyceraldehyde-3-phosphate dehydrogenase induces formation of C150–C154 intrasubunit disulfide bond in the active site of the enzyme. Biochim Biophys Acta (BBA) Gen Subj 1861:3167–3177. https://doi.org/10.1016/j.bbagen.2017.09.008
Bayles KW, Wesson CA, Liou LE, Fox LK, Bohach GA, Trumble W (1998) Intracellular staphylococcus aureus escapes the endosome and induces apoptosis in epithelial cells. Infect Immun 66:336–342
Beinert H, Kennedy MC, Stout CD (1996) Aconitase as iron− sulfur protein, enzyme, and iron-regulatory protein. Chem Rev 96:2335–2374. https://doi.org/10.1021/cr950040z
Benhar M, Stamler JS (2005) A central role for S-nitrosylation in apoptosis. Nat Cell Biol 7:645–646. https://doi.org/10.1038/ncb0705-645
Boël G, Pichereau V, Mijakovic I, Mazé A, Poncet S, Gillet S, Giard J-C, Hartke A, Auffray Y, Deutscher J (2004) Is 2-phosphoglycerate-dependent automodification of bacterial enolases implicated in their export? J Mol Biol 337:485–496. https://doi.org/10.1016/j.jmb.2003.12.082
Bogdan C (2001) Nitric oxide and the immune response. Nat Immunol 2:907–916. https://doi.org/10.1038/ni1001-907
Bose JL, Lehman MK, Fey PD, Bayles KW (2012) Contribution of the Staphylococcus aureus Atl AM and GL murein hydrolase activities in cell division, autolysis, and biofilm formation. PLoS One 7:e42244. https://doi.org/10.1371/journal.pone
Brandes N, Schmitt S, Jakob U (2009) Thiol-based redox switches in eukaryotic proteins. Antioxid Redox Signal 11:997–1014. https://doi.org/10.1089/ars.2008.2285
Bullen J, Griffiths E, Edmiston CE (1999) Iron and infection: molecular, physiological and clinical aspects. LWW. https://doi.org/10.1002/jobm.3620270810
Carpena N, Manning KA, Dokland T, Marina A, Penadés JR (2016) Convergent evolution of pathogenicity islands in helper cos phage interference. Philosoph Transactions Royal Soc B Biol Sci 371:20150505. https://doi.org/10.1098/rstb.2015.0505
Chartier FJ, Couture M (2007) Substrate-specific interactions with the heme-bound oxygen molecule of nitric-oxide synthase. J Biol Chem 282:20877–20886. https://doi.org/10.1074/jbc.m701800200
Chatterjee I, Becker P, Grundmeier M, Bischoff M, Somerville GA, Peters G, Sinha B, Harraghy N, Proctor RA, Herrmann M (2005) Staphylococcus aureus ClpC is required for stress resistance, aconitase activity, growth recovery, and death. J Bacteriol 187:4488–4496. https://doi.org/10.1128/JB.187.13.4488-4496.2005
Chen C, Zabad S, Liu H, Wang W, Jeffery CJ (2018) Moonprot 2.0: an expansion and update of the moonlighting proteins database. Nucleic Acids Res 46:640–644. https://doi.org/10.1093/nar/gkx1043
Chhatwal GS (2002) Anchorless adhesins and invasins of Gram-positive bacteria: a new class of virulence factors. Trends Microbiol 10:205–208. https://doi.org/10.1016/s0966-842x(02)02351-x
Choby JE, Mike LA, Mashruwala AA, Dutter BF, Dunman PM, Sulikowski GA, Boyd JM, Skaar EP (2016) A small-molecule inhibitor of iron-sulfur cluster assembly uncovers a link between virulence regulation and metabolism in Staphylococcus aureus. Cell Chem Biol 23:1351–1361
Coady A, Xu M, Phung Q, Cheung TK, Bakalarski C, Alexander MK, Lehar SM, Kim J, Park S, Tan M-W (2015) The Staphylococcus aureus ABC-type manganese transporter MntABC is critical for reinitiation of bacterial replication following exposure to phagocytic oxidative burst. PLoS One 10:e0138350. https://doi.org/10.1371/journal.pone
Cole N, Hume E, Khan S, Willcox M (2010) The Role of Glyceraldehyde Phosphate Dehydrogenase in the Pathogenesis of Staphylococcus aureus Keratitis. Invest Ophthalmol Vis Sci 51:3889–3889
Commichau FM, Stülke J (2008) Trigger enzymes: bifunctional proteins active in metabolism and in controlling gene expression. Mol Microbiol 67:692–702. https://doi.org/10.1111/j.1365-2958.2007.06071.x
Commichau FM, Stülke J (2015) Trigger enzymes: coordination of metabolism and virulence gene expression. Metab Bact Pathogen. https://doi.org/10.1128/microbiolspec.MBP-0010-2014
Copley SD (2012) Moonlighting is mainstream: paradigm adjustment required. BioEssays 34:578–588. https://doi.org/10.1002/bies.201100191
Corbier C, Michels S, Wonacott AJ, Branlant G (1994) Characterization of the two anion-recognition sites of glyceraldehyde-3-phosphate dehydrogenase from Bacillus stearothermophilus by site-directed mutagenesis and chemical modification. Biochemistry 33:3260–3265. https://doi.org/10.1021/bi00177a017
Cramton SE, Gerke C, Schnell NF, Nichols WW, Götz F (1999) The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect Immun 67:5427–5433
Davenport RC, Bash PA, Seaton BA, Karplus M, Petsko GA, Ringe D (1991) Structure of the triosephosphate isomerase-phosphoglycolohydroxamate complex: an analog of the intermediate on the reaction pathway. Biochemistry 30(24):5821–5826
Deivanayagam CC, Wann ER, Chen W, Carson M, Rajashankar KR, Höök M, Narayana SV (2002) A novel variant of the immunoglobulin fold in surface adhesins of Staphylococcus aureus: crystal structure of the fibrinogen-binding MSCRAMM, clumping factor A. EMBO J 21:6660–6672. https://doi.org/10.1093/emboj/cdf619
Diep BA, Gill SR, Chang RF, Phan TH, Chen JH, Davidson MG, Lin F, Lin J, Carleton HA, Mongodin EF (2006) Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet 367:731–739. https://doi.org/10.1016/S0140-6736(06)68231-7
Ebner P, Prax M, Nega M, Koch I, Dube L, Yu W, Rinker J, Popella P, Flötenmeyer M, Götz F (2015) Excretion of cytoplasmic proteins (ECP) in Staphylococcus aureus. Mol Microbiol 97:775–789. https://doi.org/10.1111/mmi.13065
Ebner P, Rinker J, Nguyen MT, Popella P, Nega M, Luqman A, Schittek B, Di Marco M, Stevanovic S, Götz F (2016) Excreted cytoplasmic proteins contribute to pathogenicity in Staphylococcus aureus. Infect Immun 84:1672–1681
Elasri M, Thomas J, Skinner R, Blevins J, Beenken K, Nelson C, Smelter M (2002) Staphylococcus aureus collagen adhesin contributes to the pathogenesis of osteomyelitis. Bone 30:275–280. https://doi.org/10.1016/S8756-3282(01)00632-9
Ellington AD, Szostak JW (1992) Selection in vitro of single-stranded DNA molecules that fold into specific ligand-binding structures. Nature 355:850–852. https://doi.org/10.1038/355850a0
Even S, Burguiere P, Auger S, Soutourina O, Danchin A, Martin-Verstraete I (2006) Global control of cysteine metabolism by CymR in Bacillus subtilis. J Bacteriol 188:2184–2197. https://doi.org/10.1128/JB.188.6.2184-2197.2006
Fitzgerald JR, Loughman A, Keane F, Brennan M, Knobel M, Higgins J, Visai L, Speziale P, Cox D, Foster TJ (2006) Fibronectin-binding proteins of Staphylococcus aureus mediate activation of human platelets via fibrinogen and fibronectin bridges to integrin GPIIb/IIIa and IgG binding to the FcγRIIa receptor. Mol Microbiol 59:212–230. https://doi.org/10.1111/j.1365-2958.2005.04922.x
Flannagan RS, Cosío G, Grinstein S (2009) Antimicrobial mechanisms of phagocytes and bacterial evasion strategies. Nat Rev Microbiol 7:355–366. https://doi.org/10.1038/nrmicro2128
Foulston L, Elsholz AK, DeFrancesco AS, Losick R (2014) The extracellular matrix of Staphylococcus aureus biofilms comprises cytoplasmic proteins that associate with the cell surface in response to decreasing pH. MBio 5:e01667–e11614. https://doi.org/10.1128/mBio.01667-14
Franco-Serrano L, Hernández S, Calvo A, Severi MA, Ferragut G, Pérez-Pons J, Piñol J, Pich Ò, Mozo-Villarias Á, Amela I, Querol E (2018a) MultitaskProtDB-II: an update of a database of multitasking/moonlighting proteins. Nucleic Acids Res 46:645–646. https://doi.org/10.1093/nar/gkx1066
Franco-Serrano L, Cedano J, Perez-Pons JA, Mozo-Villarias A, Piñol J, Amela I, Querol E (2018b) A hypothesis explaining why so many pathogen virulence proteins are moonlighting proteins. Pathog Dis. https://doi.org/10.1093/femspd/fty046(76:fty046)
Furukawa T, Yoshimura A, Sumizawa T, Haraguchi M, Akiyama S-I, Fukui K, Ishizawa M, Yamada Y (1992) Angiogenic factor. Nature 356:668–668
Furuya H, Ikeda R (2009) Interaction of triosephosphate isomerase from the cell surface of Staphylococcus aureus and α-(1→ 3)-mannooligosaccharides derived from glucuronoxylomannan of Cryptococcus neoformans. Microbiology 155:2707. https://doi.org/10.1099/mic.0.028068-0
Furuya H, Ikeda R (2011) Interaction of triosephosphate isomerase from Staphylococcus aureus with plasminogen. Microbiol Immunol 55:855–862. https://doi.org/10.1111/j.1348-0421.2011.00392.x
Geoghegan JA, Monk IR, O'Gara JP, Foster TJ (2013) Subdomains N2N3 of fibronectin binding protein A mediate Staphylococcus aureus biofilm formation and adherence to fibrinogen using distinct mechanisms. J Bacteriol 195:2675–2683. https://doi.org/10.1128/JB.02128-12
Gill SR, Fouts DE, Archer GL, Mongodin EF, DeBoy RT, Ravel J, Paulsen IT, Kolonay JF, Brinkac L, Beanan M (2005) Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J Bacteriol 187:2426–2438. https://doi.org/10.1128/JB.187.7.2426-2438.2005
Glenting J, Beck HC, Vrang A, Riemann H, Ravn P, Hansen AM, Antonsson M, Ahrné S, Israelsen H, Madsen S (2013) Anchorless surface associated glycolytic enzymes from Lactobacillus plantarum 299v bind to epithelial cells and extracellular matrix proteins. Microbiol Res 168:245–253. https://doi.org/10.1016/j.micres.2013.01.003
Goji N, Potter AA, Perez-Casal J (2004) Characterization of two proteins of Staphylococcus aureus isolated from bovine clinical mastitis with homology to glyceraldehyde-3-phosphate dehydrogenase. Vet Microbiol 99:269–279. https://doi.org/10.1016/j.vetmic.2003.12.009
Goncheva MI, Flannagan RS, Sterling BE, Laakso HA, Friedrich NC, Kaiser JC, Watson DW, Wilson CH, Sheldon JR, McGavin MJ (2019) Stress-induced inactivation of the Staphylococcus aureus purine biosynthesis repressor leads to hypervirulence. Nat Commun 10:1–14. https://doi.org/10.1038/s41467-019-08724-x
Grayczyk JP, Harvey CJ, Laczkovich I, Alonzo F III (2017) A lipoylated metabolic protein released by Staphylococcus aureus suppresses macrophage activation. Cell Host Microbe 22(678–687):e679. https://doi.org/10.1016/j.chom.2017.09.004
Greenberg EP (2000) Pump up the versatility. Nature 406:947–948. https://doi.org/10.1038/35023203
Gupta R, Bhatty M, Swiatlo E, Nanduri B (2013) Role of an iron-dependent transcriptional regulator in the pathogenesis and host response to infection with Streptococcus pneumoniae. PLoS One 8:e55157. https://doi.org/10.1371/journal.pone
Hajighahramani N, Nezafat N, Eslami M, Negahdaripour M, Rahmatabadi SS, Ghasemi Y (2017) Immunoinformatics analysis and in silico designing of a novel multi-epitope peptide vaccine against Staphylococcus aureus. Infect Gene Evol 48:83–94. https://doi.org/10.1016/j.meegid.2016.12.010
Handke LD, Hawkins JC, Miller AA, Jansen KU, Anderson AS (2013) Regulation of Staphylococcus aureus MntC expression and its role in response to oxidative stress. PloS one 8:e77874. https://doi.org/10.1371/journal.pone
Handke LD, Gribenko AV, Timofeyeva Y, Scully IL, Anderson AS (2018) MntC-dependent manganese transport is essential for Staphylococcus aureus oxidative stress resistance and virulence. mSphere 3:e00336–e1318. https://doi.org/10.1128/mSphere.00336-18
Hara MR, Agrawal N, Kim SF, Cascio MB, Fujimuro M, Ozeki Y, Takahashi M, Cheah JH, Tankou SK, Hester LD (2005) S-nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding. Nat Cell Biol 7:665–674. https://doi.org/10.1038/ncb1268
Haspel N, Ricklin D, Geisbrecht BV, Kavraki LE, Lambris JD (2008) Electrostatic contributions drive the interaction between Staphylococcus aureus protein Efb-C and its complement target C3d. Protein Sci 17:1894–1906. https://doi.org/10.1110/ps.036624.108
He C, Wilhelm SM, Pentland AP, Marmer BL, Grant GA, Eisen AZ, Goldberg GI (1989) Tissue cooperation in a proteolytic cascade activating human interstitial collagenase. Proc Natl Acad Sci 86:2632–2636. https://doi.org/10.1073/pnas.86.8.2632
Hedstrom L, Liechti G, JGoldbergR Gollapalli BD (2011) The antibiotic potential of prokaryotic IMP dehydrogenase inhibitors. Curr Med Chem 18:1909–1918
Heilmann C, Thumm G, Chhatwal GS, Hartleib J, Uekötter A, Peters G (2003) Identification and characterization of a novel autolysin (Aae) with adhesive properties from Staphylococcus epidermidis. Microbiology 149:2769–2778. https://doi.org/10.1128/IAI.73.8.4793-4802.2005
Heilmann C, Hartleib J, Hussain MS, Peters G (2005) The multifunctional Staphylococcus aureus autolysin aaa mediates adherence to immobilized fibrinogen and fibronectin. Infect Immun 73:4793–4802. https://doi.org/10.1128/IAI.73.8.4793-4802.2005
Henderson B, Martin A (2011) Bacterial virulence in the moonlight: Multitasking bacterial moonlighting proteins are virulence determinants in infectious disease. Infect Immun 79:3476–3491. https://doi.org/10.1128/IAI.00179-11
Hienz SA, Schennings T, Heimdahl A, Flock J-I (1996) Collagen binding of Staphylococcus aureus is a virulence factor in experimental endocarditis. J Infect Dis 174:83–88. https://doi.org/10.1093/infdis/174.1.83
Hill RL, Dokland T (2016) The type 2 dUTPase of bacteriophage ϕNM1 initiates mobilization of Staphylococcus aureus bovine pathogenicity island 1. J Mol Biol 428:142–152. https://doi.org/10.1016/j.jmb.2015.11.009
Hofer A, Crona M, Logan DT, Sjöberg BM (2012) DNA building blocks: keeping control of manufacture. Crit Rev Biochem Mol Biol 47:50–63. https://doi.org/10.3109/10409238.2011.630372
Houston P, Rowe SE, Pozzi C, Waters EM, O'Gara JP (2011) Essential role for the major autolysin in the fibronectin-binding protein-mediated Staphylococcus aureus biofilm phenotype. Infect Immun 79:1153–1165. https://doi.org/10.1128/IAI.00364-10
Hullo MF, Auger S, Soutourina O, Barzu O, Yvon M, Danchin A, Martin-Verstraete I (2007) Conversion of methionine to cysteine in Bacillus subtilis and its regulation. J Bacteriol 189:187–197. https://doi.org/10.1128/JB.01273-06
Hullo MF, Martin-Verstraete I, Soutourina O (2010) Complex phenotypes of a mutant inactivated for CymR, the global regulator of cysteine metabolism in Bacillus subtilis. FEMS Microbiol Lett 309:201–207. https://doi.org/10.1111/j.1574-6968.2010.02043.x
Jr Hartleib, Köhler N, Dickinson RB, Chhatwal GS, Sixma JJ, Hartford OM, Foster TJ, Peters G, Kehrel BE, Herrmann M (2000) Protein A is the von Willebrand factor binding protein on Staphylococcus aureus. Blood J Am Soc Hematol 96:2149–2156
Ikeda R, Saito F, Matsuo M, Kurokawa K, Sekimizu K, Yamaguchi M, Kawamoto S (2007) Contribution of the mannan backbone of cryptococcal glucuronoxylomannan and a glycolytic enzyme of Staphylococcus aureus to contact-mediated killing of Cryptococcus neoformans. J Bacteriol 189:4815–4826. https://doi.org/10.1128/JB.00412-07
Imber M, Huyen NTT, Pietrzyk-Brzezinska AJ, Loi VV, Hillion M, Bernhardt J, Thärichen L, Kolšek K, Saleh M, Hamilton CJ, Adrian L (2018) Protein S-bacillithiolation functions in thiol protection and redox regulation of the glyceraldehyde-3-phosphate dehydrogenase Gap in Staphylococcus aureus under hypochlorite stress. Antioxid Redox Signal 28:410–430. https://doi.org/10.1089/ars.2016.6897
Janzon L, Arvidson S (1990) The role of the delta-lysin gene (hld) in the regulation of virulence genes by the accessory gene regulator (agr) in Staphylococcus aureus. EMBO J 9:1391–1399. https://doi.org/10.1002/j.1460-2075.1990.tb08254.x
Jeffery CJ (1999) Moonlighting proteins. Trends Biochem Sci 24:8–11. https://doi.org/10.1016/S0968-0004(98)01335-8
Jongerius I, Jr Köhl, Pandey MK, Ruyken M, van Kessel KP, van Strijp JA, Rooijakkers SH (2007) Staphylococcal complement evasion by various convertase-blocking molecules. J Exp Med 204:2461–2471. https://doi.org/10.1084/jem.20070818
Jung KY, Cha JD, Lee SH, Woo WH, Do Seon L, Choi BK, Kim KJ (2001) Involvement of staphylococcal protein A and cytoskeletal actin in Staphylococcus aureus invasion of cultured human oral epithelial cells. J Med Microbiol 50:35–41. https://doi.org/10.1099/0022-1317-50-1-35
Kang M, Ko Y-P, Liang X, Ross CL, Liu Q, Murray BE, Höök M (2013) Collagen-binding microbial surface components recognizing adhesive matrix molecule (MSCRAMM) of Gram-positive bacteria inhibit complement activation via the classical pathway. J Biol Chem 288:20520–20531. https://doi.org/10.1074/jbc.M113.454462
Kaptain S, Downey WE, Tang C, Philpott C, Haile D, Orloff DG, Harford JB, Rouault TA, Klausner RD (1991) A regulated RNA binding protein also possesses aconitase activity. Proc Natl Acad Sci 88:10109–10113. https://doi.org/10.1073/pnas.88.22.10109
Kouzminova EA, Kuzminov A (2004) Chromosomal fragmentation in dUTPase-deficient mutants of Escherichia coli and its recombinational repair. Mol Microbiol 51:1279–1295. https://doi.org/10.1111/j.1365-2958.2003.03924.x
Kuroda M, Ohta T, Uchiyama I, Baba T, Yuzawa H, Kobayashi I, Cui L, Oguchi A, Aoki K-i, Nagai Y (2001) Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet 357:1225–1240. https://doi.org/10.1016/S0140-6736(00)04403-2
Lasa I, Gouin E, Goethals M, Vancompernolle K, David V, Vandekerckhove J, Cossart P (1997) Identification of two regions in the N-terminal domain of ActA involved in the actin comet tail formation by Listeria monocytogenes. EMBO J 16:1531–1540. https://doi.org/10.1093/emboj/16.7.1531
Leipuviene R, Theil EC (2007) The family of iron responsive RNA structures regulated by changes in cellular iron and oxygen. Cell Mol Life Sci 64:2945–2955. https://doi.org/10.1007/s00018-007-7198-4
Lijnen HR, Van Hoef B, De Cock F, Okada K, Ueshima S, Matsuo O, Collen D (1991) On the mechanism of fibrin-specific plasminogen activation by staphylokinase. J Biol Chem 266:11826–11832
Lindsay JA, Holden MT (2004) Staphylococcus aureus: superbug, super genome? Trends Microbiol 12:378–385. https://doi.org/10.1016/j.tim.2004.06.004
Lindsay JA, Ruzin A, Ross HF, Kurepina N, Novick RP (1998) The gene for toxic shock toxin is carried by a family of mobile pathogenicity islands in Staphylococcus aureus. Mol Microbiol 29:527–543. https://doi.org/10.1046/j.1365-2958.1998.00947.x
Lopes JD, Dos Reis M, Brentani RR (1985) Presence of laminin receptors in Staphylococcus aureus. Science 229:275–277. https://doi.org/10.1126/science.3160113
Lottenberg R, Minning-Wenz D, Boyle MD (1994) Capturing host plasmin (ogen): a common mechanism for invasive pathogens? Trends Microbiol 2:20–24. https://doi.org/10.1016/0966-842X(94)90340-9
Loughman A, Sweeney T, Keane FM, Pietrocola G, Speziale P, Foster TJ (2008) Sequence diversity in the A domain of Staphylococcus aureusfibronectin-binding protein A. BMC Microbiol 8:74. https://doi.org/10.1186/1471-2180-8-74
Lowy FD (1998) Staphylococcus aureus infections. N Engl J Med 339:520–532. https://doi.org/10.1056/NEJM199808203390806
Macdonald A, Smith A (1984) Classics in infectious diseases.” On abscesses". Alexander Ogston (1844–1929). Rev Infect Dis 6:122–128. https://doi.org/10.1093/clinids/6.1.122
Meyer-Siegler K, Mauro DJ, Seal G, Wurzer J, Deriel JK, Sirover MA (1991) A human nuclear uracil DNA glycosylase is the 37-kDa subunit of glyceraldehyde-3-phosphate dehydrogenase. Proc Natl Acad Sci 88:8460–8464. https://doi.org/10.1073/pnas.88.19.8460
Mintz KP, Fives-Taylor PM (1994) Adhesion of Actinobacillus actinomycetemcomitans to a human oral cell line. Infect Immun 62:3672–3678
Miyashita C, Wenzel E, Heiden M (1988) Plasminogen: a brief introduction into its biochemistry and function. Pathophysiol Haemost Thromb 18:7–13. https://doi.org/10.1159/000215824
Modun B, Williams P (1999) The staphylococcal transferrin-binding protein is a cell wall glyceraldehyde-3-phosphate dehydrogenase. Infect Immun 67:1086–1092. https://doi.org/10.1128/IAI.67.3.1086-1092.1999
Modun B, Evans RW, Joannou CL, Williams P (1998) Receptor-Mediated Recognition and Uptake of Iron from Human Transferrin by Staphylococcus aureus and Staphylococcus epidermidis. Infect Immun 66:3591–3596. https://doi.org/10.1128/IAI.66.8.3591-3596.1998
Mölkänen T, Tyynelä J, Helin J, Kalkkinen N, Kuusela P (2002) Enhanced activation of bound plasminogen on Staphylococcus aureus by staphylokinase. FEBS Lett 517:72–78. https://doi.org/10.1016/S0014-5793(02)02580-2
Mongodin E, Finan J, Climo MW, Rosato A, Gill S, Archer GL (2003) Microarray transcription analysis of clinical Staphylococcus aureus isolates resistant to vancomycin. J Bacteriol 185:4638–4643. https://doi.org/10.1128/JB.185.15.4638-4643.2003
Morgan E, Brown B (1977) Iron release from transferrin is mediated by organic phosphate compounds. In: Brown, Elmer B (ed) Proteins of Iron Metabolism, 1st edn. Grune and Stratton, New York, pp 227–236
Mowbray S, Koshland D (1990) Mutations in the aspartate receptor of Escherichia coli which affect aspartate binding. J Biol Chem 265:15638–15643
Muro-Pastor AM, Ostrovsky P, Maloy S (1997) Regulation of gene expression by repressor localization: biochemical evidence that membrane and DNA binding by the PutA protein are mutually exclusive. J Bacteriol 179:2788–2791. https://doi.org/10.1128/jb.179.8.2788-2791.1997
Nilsson IM, Patti JM, Bremell T, Höök M, Tarkowski A (1998) Vaccination with a recombinant fragment of collagen adhesin provides protection against Staphylococcus aureus-mediated septic death. J Clin Investig 101:2640–2649
Nordlund P, Reichard P (2006) Ribonucleotide reductases. Annu Rev Biochem 75:681–706. https://doi.org/10.1146/annurev.biochem.75.103004.142443
Novick RP (1989) Staphylococcal plasmids and their replication. Annu Rev Microbiol 43:537–563
Novick RP, Geisinger E (2008) Quorum sensing in staphylococci. Annu Rev Genet 42:541–564. https://doi.org/10.1146/annurev.genet.42.110807.091640
Ogston A (1984) On abscesses. Rev Infect Dis 6:122–128
Pancholi V, Fischetti VA (1992) A major surface protein on group A streptococci is a glyceraldehyde-3-phosphate-dehydrogenase with multiple binding activity. J Exp Med 176:415–426. https://doi.org/10.1084/jem.176.2.415
Pasztor L, Ziebandt A-K, Nega M, Schlag M, Haase S, Franz-Wachtel M, Madlung J, Nordheim A, Heinrichs DE, Götz F (2010) Staphylococcal major autolysin (Atl) is involved in excretion of cytoplasmic proteins. J Biol Chem 285:36794–36803. https://doi.org/10.1074/jbc.M110.167312
Patti JM, Bremell T, Krajewska-Pietrasik D, Abdelnour A, Tarkowski A, Rydén C, Höök M (1994) The Staphylococcus aureus collagen adhesin is a virulence determinant in experimental septic arthritis. Infect Immun 62:152–161
Penadés JR, Christie GE (2015) The phage-inducible chromosomal islands: a family of highly evolved molecular parasites. Ann Rev Virol 2:181–201. https://doi.org/10.1146/annurev-virology-031413-085446
Periasamy S, Joo H-S, Duong AC, Bach T-HL, Tan VY, Chatterjee SS, Cheung GY, Otto M (2012) How Staphylococcus aureus biofilms develop their characteristic structure. Proc Natl Acad Sci 109:1281–1286. https://doi.org/10.1073/pnas.1115006109
Piatigorsky J (1998) Multifunctional lens crystallins and corneal enzymes: more than meets the eye. Ann NY Acad Sci 842:7–15. https://doi.org/10.1111/j.1749-6632.1998.tb09626.x
Pietrocola G, Nobile G, Gianotti V, Zapotoczna M, Foster TJ, Geoghegan JA, Speziale P (2016) Molecular interactions of human plasminogen with fibronectin-binding protein B (FnBPB), a fibrinogen/fibronectin-binding protein from Staphylococcus aureus. J Biol Chem 291:18148–18162. https://doi.org/10.1074/jbc.M116.731125
Pozzi C, Waters EM, Rudkin JK, Schaeffer CR, Lohan AJ, Tong P, Loftus BJ, Pier GB, Fey PD, Massey RC (2012) Methicillin resistance alters the [VH1] biofilm phenotype and attenuates virulence in Staphylococcus aureus device-associated infections. PLoS Pathogens 8:e1002626. https://doi.org/10.1371/journal.ppat.1002626
Prasad L, Leduc Y, Hayakawa K, Delbaere LT (2004) The structure of a universally employed enzyme: V8 protease from Staphylococcus aureus. Acta Crystallogr D Biol Crystallogr 60:256–259. https://doi.org/10.1107/S090744490302599X
Purves J, Cockayne A, Moody PC, Morrissey JA (2010) Comparison of the regulation, metabolic functions, and roles in virulence of the glyceraldehyde-3-phosphate dehydrogenase homologues gapA and gapB in Staphylococcus aureus. Infect Immun 78:5223–5232. https://doi.org/10.1128/IAI.00762-10
Ralser M, Wamelink MM, Kowald A, Gerisch B, Heeren G, Struys EA, Klipp E, Jakobs C, Breitenbach M, Lehrach H, Krobitsch S (2007) Dynamic rerouting of the carbohydrate flux is key to counteracting oxidative stress. J Biol 6:10. https://doi.org/10.1186/jbiol61
Reichard P, Baldesten A, Rutberg L (1961) Formation of deoxycytidine phosphates from cytidine phosphates in extracts from Escherichia coli. J Biol Chem 236:1150–1157
Rhem MN, Lech EM, Patti JM, McDevitt D, Höök M, Jones DB, Wilhelmus KR (2000) The collagen-binding adhesin is a virulence factor in Staphylococcus aureus keratitis. Infect Immun 68:3776–3779. https://doi.org/10.1128/IAI.68.6.3776-3779.2000
Rouault TA, Klausner RD (1996) Iron-sulfur clusters as biosensors of oxidants and iron. Trends Biochem Sci 21:174–177. https://doi.org/10.1016/S0968-0004(96)10024-4
Ruffing U, Akulenko R, Bischoff M, Helms V, Herrmann M, von Müller L (2012) Matched-cohort DNA microarray diversity analysis of methicillin sensitive and methicillin resistant Staphylococcus aureus isolates from hospital admission patients. PLoS One 7:e52487. https://doi.org/10.1371/journal.pone.0052487
Salazar N, Castiblanco-Valencia MM, da Silva LB, de Castro ÍA, Monaris D, Masuda HP, Barbosa AS, Arêas APM (2014) Staphylococcus aureus manganese transport protein C (MntC) is an extracellular matrix-and plasminogen-binding protein. PLoS One 9:e112730
Schlag M, Biswas R, Krismer B, Kohler T, Zoll S, Yu W, Schwarz H, Peschel A, Götz F (2010) Role of staphylococcal wall teichoic acid in targeting the major autolysin Atl. Mol Microbiol 75:864–873. https://doi.org/10.1111/j.1365-2958.2009.07007.x
Schröder A, Kland R, Peschel A, Von Eiff C, Aepfelbacher M (2006) Live cell imaging of phagosome maturation in Staphylococcus aureus infected human endothelial cells: small colony variants are able to survive in lysosomes. Med Microbiol Immunol 195:185–194. https://doi.org/10.1007/s00430-006-0015-0
Semple JW, Italiano JE, Freedman J (2011) Platelets and the immune continuum. Nat Rev Immunol 11:264–274. https://doi.org/10.1038/nri2956
Shanks RM, Donegan NP, Graber ML, Buckingham SE, Zegans ME, Cheung AL, O'Toole GA (2005) Heparin stimulates Staphylococcus aureus biofilm formation. Infect Immun 73:4596–4606. https://doi.org/10.1128/IAI.73.8.4596-4606.2005
Shenton D, Grant CM (2003) Protein S-thiolation targets glycolysis and protein synthesis in response to oxidative stress in the yeast Saccharomyces cerevisiae. Biochem J 374:513–519. https://doi.org/10.1042/BJ20030414
Sinha B, François PP, Nüße O, Foti M, Hartford OM, Vaudaux P, Foster TJ, Lew DP, Herrmann M, Krause KH (1999) Fibronectin-binding protein acts as Staphylococcus aureus invasin via fibronectin bridging to integrin α5β1. Cell Microbiol 1:101–117. https://doi.org/10.1046/j.1462-5822.1999.00011.x
Sirover MA (2011) On the functional diversity of glyceraldehyde-3-phosphate dehydrogenase: biochemical mechanisms and regulatory control. Biochimica Biophysica Acta (BBA) Gen Subj 1810:741–751. https://doi.org/10.1016/j.bbagen.2011.05.010
Sirover MA (2012) Subcellular dynamics of multifunctional protein regulation: mechanisms of GAPDH intracellular translocation. J Cell Biochem 113:2193–2200. https://doi.org/10.1002/jcb.24113
Sirover MA (2014) Structural analysis of glyceraldehyde-3-phosphate dehydrogenase functional diversity. Int JBiochem Cell Biol 57:20–26. https://doi.org/10.1016/j.biocel.2014.09.026
Skaar EP, Schneewind O (2004) Iron-regulated surface determinants (Isd) of Staphylococcus aureus: stealing iron from heme. Microbes Infect 6:390–397. https://doi.org/10.1016/j.micinf.2003.12.008
Somerville G, Mikoryak CA, Reitzer L (1999) Physiological characterization of Pseudomonas aeruginosa during exotoxin A synthesis: glutamate, iron limitation, and aconitase activity. J Bacteriol 181:1072–1078. https://doi.org/10.1128/jb.181.4.1072-1078.1999
Somerville GA, Morgan CI, Fitzgerald JR, Dorward DW, Reitzer LJ, Musser JM (2002) Staphylococcus aureus aconitase inactivation unexpectedly inhibits post-exponential-phase growth and enhances stationary-phase survival. Infect Immun 70:6373–6382. https://doi.org/10.1128/IAI.70.11.6373-6382.2002
Stülke J (2002) Control of transcription termination in bacteria by RNA-binding proteins that modulate RNA structures. Arch Microbiol 177:433–440. https://doi.org/10.1007/s00203-002-0407-5
Suzuki CK, Rep M, Van Dijl JM, Suda K, Grivell LA, Schatz G (1997) ATP-dependent proteases that also chaperone protein biogenesis. Trends Biochem Sci 22:118–123. https://doi.org/10.1016/S0968-0004(97)01020-7
Tanous C, Soutourina O, Raynal B, Hullo MF, Mervelet P, Gilles AM, Noirot P, Danchin A, England P, Martin-Verstraete I (2008) The CymR regulator in complex with the enzyme CysK controls cysteine metabolism in Bacillus subtilis. J Biol Chem 283:35551–35560. https://doi.org/10.1074/jbc.M805951200Epub 2008 Oct 29
Taylor JM, Heinrichs DE (2002) Transferrin binding in Staphylococcus aureus: involvement of a cell wall-anchored protein. Mol Microbiol 43:1603–1614. https://doi.org/10.1046/j.1365-2958.2002.02850.x
Tormo-Más MÁ, Mir I, Shrestha A, Tallent SM, Campoy S, Lasa Í, Barbé J, Novick RP, Christie GE, Penadés JR (2010) Moonlighting bacteriophage proteins derepress staphylococcal pathogenicity islands. Nature 465:779–782. https://doi.org/10.1038/nature09065
Tormo-Más MÁ, Donderis J, García-Caballer M, Alt A, Mir-Sanchis I, Marina A, Penadés JR (2013) Phage dUTPases control transfer of virulence genes by a proto-oncogenic G protein-like mechanism. Mol Cell 49:947–958. https://doi.org/10.1016/j.molcel.2012.12.013
Tristan C, Shahani N, Sedlak TW, Sawa A (2011) The diverse functions of GAPDH: views from different subcellular compartments. Cell Signal 23:317–323. https://doi.org/10.1016/j.cellsig.2010.08.003
Tsuchiya Y, Zhyvoloup A, Baković J, Thomas N, Yu BYK, Das S, Orengo C, Newell C, Ward J, Saladino G, Comitani F (2018) Protein CoAlation and antioxidant function of coenzyme A in prokaryotic cells. Biochem J 475:1909–1937. https://doi.org/10.1042/bcj20180043
Valotteau C, Prystopiuk V, Pietrocola G, Rindi S, Peterle D, De Filippis V, Foster TJ, Speziale P, Dufrêne YF (2017) Single-cell and single-molecule analysis unravels the multifunctionality of the Staphylococcus aureus collagen-binding protein Cna. ACS Nano 11:2160–2170. https://doi.org/10.1021/acsnano.6b08404
Van der Ploeg JR, Barone M, Leisinger T (2001) Functional analysis of the Bacillus subtilis cysK and cysJI genes. FEMS Microbiol Lett 201:29–35. https://doi.org/10.1111/j.1574-6968.2001.tb10728.x
Van Laer K, Hamilton CJ, Messens J (2013) Low-molecular-weight thiols in thiol–disulfide exchange. Antioxid Redox Signal 18:1642–1653. https://doi.org/10.1089/ars.2012.4964
Vértessy BG, Tóth J (2009) Keeping uracil out of DNA: physiological role, structure and catalytic mechanism of dUTPases. Acc Chem Res 42:97–106. https://doi.org/10.1021/ar800114w
Volz K (2008) The functional duality of iron regulatory protein 1. Curr Opin Struct Biol 18:106–111. https://doi.org/10.1016/j.sbi.2007.12.010
Walden WE, Selezneva AI, Dupuy J, Volbeda A, Fontecilla-Camps JC, Theil EC, Volz K (2006) Structure of dual function iron regulatory protein 1 complexed with ferritin IRE-RNA. Science 314:1903–1908. https://doi.org/10.1126/science.1133116
Walport MJ (2001) Complement. N Engl J Med 344:1058–1066
Weber H, Engelmann S, Becher D, Hecker M (2004) Oxidative stress triggers thiol oxidation in the glyceraldehyde-3-phosphate dehydrogenase of Staphylococcus aureus. Mol Microbiol 52:133–140. https://doi.org/10.1111/j.1365-2958.2004.03971.x
Wegrzyn G (2005) What does “plasmid biology” currently mean?: summary of the Plasmid Biology 2004 meeting. Plasmid 53:14–22. https://doi.org/10.1016/j.plasmid.2004.10.002
Weinberg ED (1978) Iron and infection. Microbiol Rev 42:45
Wold F (1971) Enolase. In: Boyer PD (ed) The Enzymes, 3rd edn. Elsevier B.V, Netherlands, pp 499–538. https://doi.org/10.1016/S1874-6047(08)60101-8
Wu Y, Wang C, Lin S, Wu M, Han L, Tian C, Zhang X, Zang J (2015) Octameric structure of Staphylococcus aureus enolase in complex with phosphoenolpyruvate. Acta Crystallogr D Biol Crystallogr 71:2457–2470. https://doi.org/10.1107/S1399004715018830/qh5032sup1.pdf
Yamada S, Sugai M, Komatsuzawa H, Nakashima S, Oshida T, Matsumoto A, Suginaka H (1996) An autolysin ring associated with cell separation of Staphylococcus aureus. J Bacteriol 178:1565–1571. https://doi.org/10.1128/jb.178.6.1565-1571.1996
Zaffagnini M, Michelet L, Marchand C, Sparla F, Decottignies P, Le Maréchal P, Miginiac-Maslow M, Noctor G, Trost P, Lemaire SD (2007) The thioredoxin-independent isoform of chloroplastic glyceraldehyde-3-phosphate dehydrogenase is selectively regulated by glutathionylation. FEBS J 274:212–226. https://doi.org/10.1111/j.1742-4658.2006.05577.x
Zhang R-g, Evans G, Rotella FJ, Westbrook EM, Beno D, Huberman E, Joachimiak A, Collart FR (1999) Characteristics and crystal structure of bacterial inosine-5 ‘-monophosphate dehydrogenase. Biochemistry 38:4691–4700. https://doi.org/10.1021/bi982858v
Zhang H, Luo Q, Gao H, Feng Y (2015) A new regulatory mechanism for bacterial lipoic acid synthesis. Microbiology Open 4:282–300. https://doi.org/10.1002/mbo3.237
Zhao C, Moriga Y, Feng B, Kumada Y, Imanaka H, Imamura K, Nakanishi K (2006) On the interaction site of serine acetyltransferase in the cysteine synthase complex from Escherichia coli. Biochem Biophys Res Commun 341:911–916. https://doi.org/10.1016/j.bbrc.2006.01.054
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Erko stackebrandt.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Hemmadi, V., Biswas, M. An overview of moonlighting proteins in Staphylococcus aureus infection. Arch Microbiol 203, 481–498 (2021). https://doi.org/10.1007/s00203-020-02071-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-020-02071-y