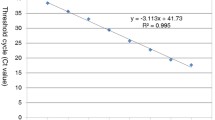
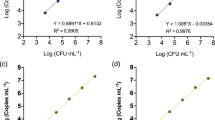
Abstract
Azospirillum brasilense is a plant growth promoting bacteria used as an inoculant in diverse crops. Accurate analytical methods are required to enumerate viable cells in inoculant formulations or in planta. We developed a quantitative polymerase chain reaction (qPCR) assay associated to propidium monoazide (PMA) to evaluate the cell viability of A. brasilense in inoculant and in maize roots. A. brasilense was grown in culture medium and was exposed to 50 ℃. Maize roots were grown in vitro and harvested 7 days after inoculation. Quantification was performed by qPCR, PMA-qPCR, and plate counting. Standard curves efficiency values ranged from 85 to 99%. The limit of detection was 104 CFU per gram of fresh root. Enumeration obtained in maize roots by qPCR where higher than enumeration by PMA-qPCR and by plate counting. PMA-qPCR assay was efficient in quantifying inoculant viable cells and provides reliable results in a quickly and accurately way compared to culture-dependent methods.




Similar content being viewed by others
References
Al-Daoud F, Gossen BD, Robson J, McDonald MR (2017) Propidium monoazide improves quantification of resting spores of Plasmodiophora brassicae with qPCR plant disease. Plant Dis 101:442–447
Balsanelli E et al (2010) Herbaspirillum seropedicae rfbB and rfbC genes are required for maize colonization. Environ Microbiol 12:2233–2244. https://doi.org/10.1111/j.1462-2920.2010.02187.x
Bashan Y, Holguin G, De-Bashan LE (2004) Azospirillum-plant relationships: physiological, molecular, agricultural, and environmental advances (1997–2003). Can J Microbiol 50:521–577
Berninger T, González López Ó, Bejarano A, Preininger C, Sessitsch A (2017) Maintenance and assessment of cell viability in formulation of non-sporulating bacterial inoculants. Microb Biotechnol 11:277–301. https://doi.org/10.1111/1751-7915.12880
Bustin SA et al (2009) The MIQE guidelines: minimumiInformation for publication of quantitative real-time PCR experiments. Clin Chem 55:611–622. https://doi.org/10.1373/clinchem.2008.112797
Couillerot O, Bouffaud M-L, Baudoin E, Muller D, Caballero-Mellado J, Moënne-Loccoz Y (2010) Development of a real-time PCR method to quantify the PGPR strain Azospirillum lipoferum CRT1 on maize seedlings. Soil Biol Biochem 42:2298–2305. https://doi.org/10.1016/j.soilbio.2010.09.003
Crespo-Sempere A, Estiarte N, Marín S, Sanchis V, Ramos AJ (2013) Propidium monoazide combined with real-time quantitative PCR to quantify viable Alternaria spp. contamination in tomato products. Int J Food Microbiol 165:214–220
Daranas N, Bonaterra A, Francés J, Cabrefiga J, Montesinos E, Badosa E (2018) Monitoring viable cells of the biological control agent Lactobacillus plantarum PM411 in aerial plant surfaces by means of a strain-specific viability quantitative PCR method. Appl Environ Microbiol 84:e00107–18
Dinu L-D, Bach S (2013) Detection of viable but non-culturable Escherichia coli O157: H7 from vegetable samples using quantitative PCR with propidium monoazide and immunological assays. Food Control 31:268–273
Dobbelaere S, Vanderleyden J, Okon Y (2003) Plant growth-promoting effects of diazotrophs in the rhizosphere. Crit Rev Plant Sci 22:107–149
Egener T, Hurek T, Reinhold-Hurek B (1999) Endophytic expression of nif genes of Azoarcus sp. Strain BH72 in Rice. Roots Mol Plant Microbe Interact 12:813–819. https://doi.org/10.1094/MPMI.1999.12.9.813
Elizaquível P, Sánchez G, Aznar R (2012) Quantitative detection of viable foodborne E. coli O157: H7, Listeria monocytogenes and Salmonella in fresh-cut vegetables combining propidium monoazide and real-time PCR. Food Control 25:704–708
Faleiro AC, Pereira TP, Espindula E, Brod FCA, Arisi ACM (2013) Real time PCR detection targeting nifA gene of plant growth promoting bacteria Azospirillum brasilense strain FP2 in maize roots. Symbiosis 61:125–133. https://doi.org/10.1007/s13199-013-0262-y
Fibach-Paldi S, Burdman S, Okon Y (2012) Key physiological properties contributing to rhizosphere adaptation and plant growth promotion abilities of Azospirillum brasilense. FEMS Microbiol Lett 326:99–108
Fittipaldi M, Nocker A, Codony F (2012) Progress in understanding preferential detection of live cells using viability dyes in combination with DNA amplification. J Microbiol Methods 91:276–289. https://doi.org/10.1016/j.mimet.2012.08.007
Garcia-Cayuela T, Tabasco R, Pelaez C, Requena T (2009) Simultaneous detection and enumeration of viable lactic acid bacteria and bifidobacteria in fermented milk by using propidium monoazide and real-time PCR. Int Dairy J 19:405–409. https://doi.org/10.1016/j.idairyj.2009.02.001
Gensberger ET, Polt M, Konrad-Köszler M, Kinner P, Sessitsch A, Kostić T (2014) Evaluation of quantitative PCR combined with PMA treatment for molecular assessment of microbial water quality. Water Res 67:367–376
Gómez-Rojo EM, Romero-Santacreu L, Jaime I, Rovira J (2015) A novel real-time PCR assay for the specific identification and quantification of Weissella viridescens in blood sausages. Int J Food Microbiol 215:16–24. https://doi.org/10.1016/j.ijfoodmicro.2015.08.002
Hougs L et al. (2017) Verification of analytical methods for GMO testing when implementing interlaboratory validated methods. EUR 29015 EN, Publication Office of the European Union. JRC 109940. https://doi.org/10.2760/645114
Hungria M, Campo RJ, Souza EM, Pedrosa FO (2010) Inoculation with selected strains of Azospirillum brasilense and A. lipoferum improves yields of maize and wheat in Brazil. Plant and Soil 331:413–425. https://doi.org/10.1007/s11104-009-0262-0
Ilha EC, Scariot MC, Treml D, Pereira TP, Sant′Anna ES, Prudêncio ES, Arisi ACM (2016) Comparison of real-time PCR assay and plate count for Lactobacillus paracasei enumeration in yoghurt. Ann Microbiol 66:597–606. https://doi.org/10.1007/s13213-015-1137-7
Khare E, Arora NK (2011) Physiologically stressed cells of fluorescent Pseudomonas EKi as better option for bioformulation development for management of charcoal rot caused by Macrophomina phaseolina in field conditions. Curr Microbiol 62:1789
Li F et al (2016) Rapid and simultaneous detection of viable Cronobacter sakazakii, Staphylococcus aureus, and Bacillus cereus in infant food products by PMA-mPCR assay with internal amplification control LWT-Food. Sci Technol 74:176–182
Machado HB, Funayama S, Rigo LU, Pedrosa FO (1991) Excretion of ammonium by Azospirillum brasilense mutants resistant to ethylenediamine. Can J Microbiol 37(7):549–553. https://doi.org/10.1139/m91-092
Nkuipou-Kenfack E, Engel H, Fakih S, Nocker A (2013) Improving efficiency of viability-PCR for selective detection of live cells. J Microbiol Methods 93:20–24
Nocker A, Camper AK (2006) Selective removal of DNA from dead cells of mixed bacterial communities by use of ethidium monoazide. Appl Environ Microbiol 72:1997
Nocker A, Camper AK (2009) Novel approaches toward preferential detection of viable cells using nucleic acid amplification techniques. FEMS Microbiol Lett 291:137–142. https://doi.org/10.1111/j.1574-6968.2008.01429.x
Pedrosa FO, Yates MG (1984) Regulation of nitrogen fixation (nif) genes of Azospirillum brasilense by nifA and ntr (gln) type gene products FEMS. Microbiol Lett 23:95–101
Pereira TP, do Amaral FP, Dall’Asta P, Brod FCA, Arisi ACM (2014) Real-time PCR quantification of the plant growth promoting bacteria Herbaspirillum seropedicae strain SmR1 in Maize. Roots Mol Biotechnol 56:660–670. https://doi.org/10.1007/s12033-014-9742-4
Pribylova R, Kubickova L, Babak V, Pavlik I, Kralik P (2012) Effect of short-and long-term antibiotic exposure on the viability of Mycobacterium avium subsp. paratuberculosis as measured by propidium monoazide F57 real time quantitative PCR and culture. Vet J 194:354–360
Roncato-Maccari LDB et al (2006) Endophytic Herbaspirillum seropedicae expresses nif genes in gramineous plants. FEMS Microbiol Ecol 45:39–47. https://doi.org/10.1016/S0168-6496(03)00108-9
Scariot MC, Venturelli GL, Prudêncio ES, Arisi ACM (2018) Quantification of Lactobacillus paracasei viable cells in probiotic yoghurt by propidium monoazide combined with quantitative PCR. Int J Food Microbiol 264:1–7
Shime-Hattori A, Kobayashi S, Ikeda S, Asano R, Shime H, Shinano T (2011) A rapid and simple PCR method for identifying isolates of the genus Azospirillum within populations of rhizosphere bacteria. J Appl Microbiol 111:915–924
Soto-Muñoz L, Torres R, Usall J, Viñas I, Solsona C, Teixidó N (2015) DNA-based methodologies for the quantification of live and dead cells in formulated biocontrol products based on Pantoea agglomerans CPA-2. Int J Food Microbiol 210:79–83
Stets MI, Alqueres SMC, Souza EM, Pedrosa FdO, Schmid M, Hartmann A, Cruz LM (2015) Quantification of Azospirillum brasilense FP2 bacteria in wheat roots by strain-specific quantitative PCR. Appl Environ Microbiol 81:6700–6709. https://doi.org/10.1128/AEM.01351-15
Temple TN, du Toit LJ, Derie ML, Johnson KB (2013) Quantitative molecular detection of Xanthomonas hortorum pv. carotae in carrot seed before and after hot-water treatment. Plant Dis 97:1585–1592
Truchado P, Gil MI, Kostic T, Allende A (2016) Optimization and validation of a PMA qPCR method for Escherichia coli quantification in primary production. Food Control 62:150–156. https://doi.org/10.1016/j.foodcont.2015.10.014
Vondrakova L, Turonova H, Scholtz V, Pazlarova J, Demnerova K (2018) Impact of various killing methods on EMA/PMA-qPCR efficacy. Food Control 85:23–28
Wagner AO, Praeg N, Reitschuler C, Illmer P (2015) Effect of DNA extraction procedure, repeated extraction and ethidium monoazide (EMA)/propidium monoazide (PMA) treatment on overall DNA yield and impact on microbial fingerprints for bacteria, fungi and archaea in a reference soil. Appl Soil Ecol 93:56–64
Wisniewski-Dyé F et al (2011) Azospirillum genomes reveal transition of bacteria from aquatic to terrestrial environments. PLOS Genet 7:e1002430. https://doi.org/10.1371/journal.pgen.1002430
Yang X, Badoni M, Gill CO (2011) Use of propidium monoazide and quantitative PCR for differentiation of viable Escherichia coli from E. coli killed by mild or pasteurizing heat treatments. Food Microbiol 28:1478–1482
Yang Y et al (2013) Magnetic nano-beads based separation combined with propidium monoazide treatment and multiplex PCR assay for simultaneous detection of viable Salmonella typhimurium, Escherichia coli O157: H7 and Listeria monocytogenes in food products. Food Microbiol 34:418–424
Zhang T, Fang HHP (2006) Applications of real-time polymerase chain reaction for quantification of microorganisms in environmental samples. Appl Microbiol Biotechnol 70:281–289
Acknowledgements
We would like to express our gratitude to Emanuel Maltempi de Souza and Roseli Prado, Universidade Federal do Paraná (UFPR) for providing A. brasilense strain FP2 and H. seropedicae strain SmR1.
Funding
This study was funded by National Council of Scientific and Technological Development (CNPq), Ministry of Science and Technology, Brazil. Elisandra T. Cunha, Ana M. Pedrolo and Mirella C. Scariot are recipients of PhD fellowships Coordination of Personnel Improvement of Higher Education (CAPES), Ministry of Education, Brazil. Franciele Paludo is recipient of Master fellowship from CNPq and Ana Carolina M. Arisi is recipient of research fellowship (PQ2) from CNPq.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors declare that they have no conflict of interest.
Ethical standards
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Communicated by Erko Stackebrandt.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
da Cunha, E.T., Pedrolo, A.M., Paludo, F. et al. Azospirillum brasilense viable cells enumeration using propidium monoazide-quantitative PCR. Arch Microbiol 202, 1653–1662 (2020). https://doi.org/10.1007/s00203-020-01877-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-020-01877-0