Abstract
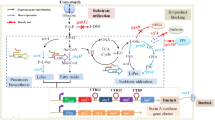
The catalytic fraction of the Cellulomonas flavigena PN-120 oligomeric β-glucosidase (BGLA) was expressed both intra- and extracellularly in a recombinant diploid of Saccharomyces cerevisiae, under limited nutrient conditions. The recombinant enzyme (BGLA15) expressed in the supernatant of a rich medium showed 582 IU/L and 99.4 IU/g dry cell, with p-nitrophenyl-β-d-glucopyranoside as substrate. BGLA15 displayed activity against cello-oligosaccharides with 2–5 glucose monomers, demonstrating that the protein is not specific for cellobiose and that the oligomeric structure is not essential for β-d-1,4-bond hydrolysis. Native β-glucosidase is inhibited almost completely at 160 mM glucose, thus limiting cellobiose hydrolysis. At 200 mM glucose concentration, BGLA15 retained more than 50 % of its maximal activity, and even at 500 mM glucose concentration, more than 30 % of its activity was preserved. Due to these characteristics of BGLA15 activity, recombinant S. cerevisiae is able to utilize cellulosic materials (cello-oligosaccharides) to produce bioethanol.





Similar content being viewed by others
References
Barrera-Islas GA, Ramos-Valdivia AC, Salgado LM, Ponce-Noyola T (2007) Characterization of a β-glucosidase produced by a high-specific growth-rate mutant of Cellulomonas flavigena. Curr Microbiol 54:266–270
Bayer EA, Raphael LR, Himmel ME (2007) The potential of cellulases and cellulosomes for cellulosic waste management. Curr Opin Biotech 18:237–245
Benoliel B, Poças-Fonseca MJ, Gonçalves Torres FA, Pepe de Moraes LM (2010) Expression of a glucose-tolerant β-glucosidase from Humicola grisea var. thermoidea in Saccharomyces cerevisiae. Appl Biochem Biotechnol 160:2036–2044
Bhatia Y, Mishra S, Bisaria VS (2002) Microbial β-glucosidases: cloning, properties and applications. Crit Rev Biotechnol 22:375–407
Chirico WI, Brown RD Jr (1985) Separation of [1-3H] cellooligosaccharides by thin-layer chromatography assay for cellulolytic enzymes. Anal Biochem 150:264–272
Cho KM, Yoo YJ, Kanq HS (1999) δ-integration of endo/exo-glucanase and β-glucosidase genes into the yeast chromosomes for direct conversion of cellulose to ethanol. Enzyme Microb Technol 25:23–30
Choi ES, Sohn JH, Rhee SK (1994) Optimization of the expression system using galactose-inducible promoter for the production of anticoagulant hirudin in Saccharomyces cerevisiae. Appl Microbiol Biotechnol 42:587–594
Den Haan R, McBride JE, La Grange DC, Lynd LR, Van Zyl W (2007a) Functional expression of cellobiohydrolases in Saccharomyces cerevisiae towards one-step conversion of cellulose to ethanol. Enzyme Microb Technol 40:1291–1299
Den Haan R, Rose SH, Lynd LR, van Zyl WH (2007b) Hydrolysis and fermentation of amorphous cellulose by recombinant S. cerevisiae. Metab Eng 9:87–94
Fujita Y, Takahashi S, Ueda M, Tanaka A, Okada H, Morikawa Y, Kawaguchi T, Arai M, Fukuda H, Kondo A (2002) Direct and efficient production of ethanol from cellulosic material with a yeast strain displaying cellulolytic enzymes. Appl Environ Microbiol 68:5136–5141
Gietz D, Woods RA (2006) Methods in molecular biology. In: Xiao Wei (ed) Yeast protocols, 2nd edn. Human Press Inc, Totowa, New Jersey, pp 107–120
Görgens JF, Passoth V, van Zyl WH, Knoetze JH, Hahn-Hägendal B (2005) Amino acids supplementation controlled oxygen limitation and sequential double induction improves heterologous xylanase production by P. stipites. FEMS Yeast Res 5:677–683
Hahn-Hägerdal B, Karhumaa K, Larsson CU, Gorwa-Grauslund M, Görgens J, van Zyl W (2005) Role of cultivation media in the development of yeast strain for large scale industrial use. Microb Cell Fact 4:1–16
Henrrissat B, Claeyssens M, Tomme P, Lemesle L, Mornon JP (1989) Cellulase families revealed by hydrophobic cluster-analysis. Gene 81:83–95
Hong J, Tamaki H, Kumagai H (2007) Cloning and functional expression of thermostable β-glucosidase gene from Thermoascus aurantiacus. Appl Microbiol Biotechnol 73:1331–1339
Hrmova M, Fichner BG (2007) Dissecting the catalytic mechanism of a plant β-D-glucan glucohydrolase through structural biology using inhibitors and substrate analogues. Carbohydr Res 342:1613–1623
Hrmova M, Streltson AV, Smith JB, Vasella A, Varghese NJ, Fincher BG (2005) Structural rationale for low-nanomolar binding of transition state mimics to a family GH3 β-D-glucan glucohydrolase from barley. Biochemistry 44:16529–16539
Lynd LR, Weimer PJ, van Zyl WH, Protorius IS (2002) Microbial cellulase utilization: fundamentals and biotechnology. Microbiol Mol Biol 66:506–577
Njokweni AP, Rose SH, van Zyl WH (2012) Fungal β-glucosidase expression in Saccharomyces cerevisiae. J Ind Microbiol Biotechnol 39:1445–1452
Pack SP, Park K, Yoo YJ (2002) Enhancement of β-glucosidase stability and cellobiose-usage using surface engineered recombinant Saccharomyces cerevisiae in ethanol production. Biotechnol Lett 24:1919–1925
Penttilä ME, Andre L, Lehtovaara P, Bailey M, Teeri TT, Knowles JK (1988) Efficient secretion of two fungal cellobiohydrolases by Saccharomyces cerevisiae. Gene 63:103–112
Ponce-Noyola T, de la Torre M (1995) Isolation of a high-specific-growth-rate mutant of Cellulomonas flavigena on sugar cane bagasse. Appl Microbiol Biotechnol 42:709–712
Ponce-Noyola T, de la Torre M (2001) Regulation of cellulases and xylanases from a derepressed mutant of Cellulomonas flavigena growing on sugar-cane bagasse in continuous culture. Bioresour Technol 78:285–291
Pozzo T, Pasten LJ, Karlsson NE, Logan DT (2010) Structural and functional analyses of β-glucosidase 3B Thermotoga neapolitana: a thermostable three-domain representative of glycoside hydrolase 3. J Mol Biol 397:724–739
Sambrook JE, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual. Cold Spring Harbor, New York
Sánchez-Torres P, González-Candelas L, Ramón D (1998) Heterologous expression of a Candida molischiana anthocyanin-β-glucosidase in a wine yeast strain. J Agric Food Chem 46:354–360
Schuster BG, Chinn MS (2012) Consolidated bioprocessing of lignocellulose feedstocks for ethanol fuel production. Bioenerg Res 6:416–435
Shen Y, Zhang Y, Ma T, Bao X, Du F, Zhuang G, Qu Y (2008) Simultaneous saccharification and fermentation of acid-pretreated corncobs with a recombinant Saccharomyces cerevisiae expressing β-glucosidase. Bioresour Technol 99:5099–5103
Skory CD, Freer SN, Bothast RJ (1996) Expression and secretion of the Candida wickerhamii extracellular β-glucosidase gene, bglB, in Saccharomyces cerevisiae. Curr Genet 30:417–422
Spiridonov NA, Wilson DB (2001) Cloning and biochemical characterization of BGLC, a β-glucosidase from the cellulolytic actinomycete Thermobifida fusca. Curr Microbiol 42:295–301
Van Zyl WH, Lynd LR, Den Haan R, Mc Bride JE (2007) Consolidate bioprocessing for bioethanol production using Saccharomyces cerevisiae. Adv Biochem Engin Biotechnol 108:205–235
Wildt S, Gerngross TU (2005) The humanization of N-glycosylation pathways in yeast. Nat Rev Microbiol 3:119–127
Yamada R, Tanaka T, Ogino C (2010) Gene copy number and polyploid on products formation in yeast. Appl Microbiol Biotechnol 88:849–857
Yamada R, Taniguchi N, Tanaka T, Ogino C, Fukuda H, Kondo A (2011) Direct ethanol production from cellulosic materials using a diploid strain of Saccharomyces cerevisiae with optimized cellulase expression. Biotechnol Biofuels 4:1–8
Yan T, Lin Y, Lin C (1998) Purification and characterization of an extracellular β-glucosidase II with high hydrolysis and transglucosylation activities from Asperillus niger. J Agric Food Chem 46:431–437
Yoshida E, Hidaka M, Foshinobu S, Koyanagi T, Minami H, Tamakis H, Kitaoka M, Katayama T, Kumagi H (2010) Role of a PA14 domain in determining substrate specificity of a glucoside hydrolase family 3 β-glucosidase from Kluyveromyces marxianus. Biochem J 431:39–49
Acknowledgments
This work was supported by Consejo Nacional de Ciencia y Tecnología México (CONACYT) (Grant 104333). D. J. Mendoza-Aguayo received a scholarship number 204305 from CONACYT México. María Isabel Pérez- Montfort corrected the English version of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Erko Stackebrandt.
Rights and permissions
About this article
Cite this article
Mendoza-Aguayo, D.J., Poggi-Varaldo, H.M., García-Mena, J. et al. Extracellular expression of glucose inhibition-resistant Cellulomonas flavigena PN-120 β-glucosidase by a diploid strain of Saccharomyces cerevisiae . Arch Microbiol 196, 25–33 (2014). https://doi.org/10.1007/s00203-013-0935-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-013-0935-1