Abstract
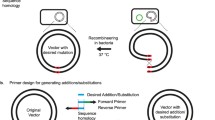
MrpA is the multimer resolution protein of the Streptomyces coelicolor A3(2) plasmid SCP2*. Previously, MrpA was found to be a site-specific tyrosine recombinase that acts with the 36-bp recombination site mrpS. The present report gives a comprehensive characterization of the composition as well as the position of the spacer and MrpA binding sites within mrpS. Experiments revealed a spacer consisting of 6 remarkably variable nucleotides in the middle of the mrpS-site. A reduction in the spacer to 5 nucleotides abolished recombination. Investigation of the MrpA binding sites showed the importance of its 15 nucleotides on an effective recombination. Among almost randomly exchangeable nucleotides, two nucleotides were identified as essential for MrpA binding. Alteration of either of these nucleotides led to a reduction in MrpA binding down to 2 % or even to no binding. Based on these results, a new left element/right element (LE/RE) deletion system was developed. The constructed heteromeric mrpS-sites are efficiently resolved by MrpA. The resulting double mutated (LE/RE) site can no longer be used as a recombination site by MrpA. The system has been successfully applied for the generation of multiple-targeted deletions in the genome of E. coli.










Similar content being viewed by others
References
Abremski K, Hoess R (1984) Bacteriophage P1 site-specific recombination. Purification and properties of the Cre recombinase protein. J Biol Chem 259:1509–1514
Araki K, Araki M, Yamamura K (1997) Targeted integration of DNA using mutant lox sites in embryonic stem cells. Nucleic Acids Res 25:868–872
Araki K, Okada Y, Araki M, Yamamura K (2010) Comparative analysis of right element mutant lox sites on recombination efficiency in embryonic stem cells. BMC Biotechnol 10:29
Baich A, Pierson DJ (1965) Control of proline synthesis in Escherichia coli. Biochim Biophys Acta 104:397–404
Banerjee A, Biswas I (2008) Markerless multiple-gene-deletion system for Streptococcus mutans. Appl Environ Microbiol 74:2037–2042
Barre FX, Soballe B, Michel B, Aroyo M, Robertson M, Sherratt D (2001) Circles: the replication-recombination-chromosome segregation connection. Proc Natl Acad Sci U S A 98:8189–8195
Bibb MJ, Hopwood DA (1981) Genetic studies of the fertility plasmid SCP2 and its SCP2* variants in Streptomyces coelicolor A3(2). J Gen Microbiol 126:427–442
Bibb MJ, Freeman RF, Hopwood DA (1977) Physical and genetical characterisation of a second sex factor, SCP2, for Streptomyces coelicolor A3(2). Mol Gen Genet 154:155–166
Bibb M, Schottel JL, Cohen SN (1980) A DNA cloning system for interspecies gene transfer in antibiotic-producing Streptomyces. Nature 284:526–531
Broach JR, Guarascio VR, Jayaram M (1982) Recombination within the yeast plasmid 2 μm circle is site-specific. Cell 29:227–234
Brown WRA, Lee NCO, Xu Z, Smith MCM (2011) Serine recombinases as tools for genome engineering. Methods 53:372–379
Carter Z, Delneri D (2010) New generation of loxP-mutated deletion cassettes for the genetic manipulation of yeast natural isolates. Yeast 27:765–775
Chang AC, Cohen SN (1978) Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol 134:1141–1156
Chung CT, Niemela SL, Miller RH (1989) One-step preparation of competent Escherichia coli: transformation and storage of bacterial cells in the same solution. Proc Natl Acad Sci U S A 86:2172–2175
Cox MM, Goodman MF, Kreuzer KN, Sherratt DJ, Sandler SJ, Marians KJ (2000) The importance of repairing stalled replication forks. Nature 404:37–41
Datsenko KA, Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645
Dubeau M-P, Ghinet MG, Jacques P-E, Clermont N, Beaulieu C, Brzezinski R (2009) Cytosine deaminase as a negative selection marker for gene disruption and replacement in the genus Streptomyces and other actinobacteria. Appl Environ Microbiol 75:1211–1214
Esposito D, Scocca JJ (1997) The integrase family of tyrosine recombinases: evolution of a conserved active site domain. Nucleic Acids Res 25:3605–3614
Fowler AV, Zabin I (1978) Amino acid sequence of beta-galactosidase. XI. Peptide ordering procedures and the complete sequence. J Biol Chem 253:5521–5525
Fried M, Crothers DM (1981) Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res 9:6505–6525
Garner MM, Revzin A (1981) A gel electrophoresis method for quantifying the binding of proteins to specific DNA regions: application to components of the Escherichia coli lactose operon regulatory system. Nucleic Acids Res 9:3047–3060
Ghosh K, Van Duyne GD (2002) Cre-loxP biochemistry. Methods 28:374–383
Grainge I, Jayaram M (1999) The integrase family of recombinase: organization and function of the active site. Mol Microbiol 33:449–456
Grainge I, Sherratt DJ (2007) Site-specific recombination. In: Aguilera A, Rothstein R (eds) Molecular genetics of recombination. Springer, Berlin, pp 443–463
Grindley NDF, Whiteson KL, Rice PA (2006) Mechanisms of site-specific recombination. Annu Rev Biochem 75:567–605
Groth AC, Calos MP (2004) Phage integrases: biology and applications. J Mol Biol 335:667–678
Guo F, Gopaul DN, van Duyne GD (1997) Structure of Cre recombinase complexed with DNA in a site-specific recombination synapse. Nature 389:40–46
Gust B, Challis GL, Fowler K, Kieser T, Chater KF (2003) PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc Natl Acad Sci U S A 100:1541–1546
Hartung M, Kisters-Woike B (1998) Cre mutants with altered DNA binding properties. J Biol Chem 273:22884–22891
Haug I, Weissenborn A, Brolle D, Bentley S, Kieser T, Altenbuchner J (2003) Streptomyces coelicolor A3(2) plasmid SCP2*: deductions from the complete sequence. Microbiology 149:505–513
Hirano N, Muroi T, Takahashi H, Haruki M (2011) Site-specific recombinases as tools for heterologous gene integration. Appl Microbiol Biotechnol 92:227–239
Hoess R, Abremski K, Irwin S, Kendall M, Mack A (1990) DNA specificity of the Cre recombinase resides in the 25 kDa carboxyl domain of the protein. J Mol Biol 216:873–882
Jeske M, Altenbuchner J (2010) The Escherichia coli rhamnose promoter rhaP(BAD) is in Pseudomonas putida KT2440 independent of Crp-cAMP activation. Appl Microbiol Biotechnol 85:1923–1933
Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM, Peterson KM (1995) Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176
Kwon HJ, Tirumalai R, Landy A, Ellenberger T (1997) Flexibility in DNA recombination: structure of the lambda integrase catalytic core. Science 276:126–131
Lee J, Jayaram M (1995) Role of partner homology in DNA recombination. Complementary base pairing orients the 5′-hydroxyl for strand joining during Flp site-specific recombination. J Biol Chem 270:4042–4052
Link AJ, Phillips D, Church GM (1997) Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J Bacteriol 179:6228–6237
Lydiate DJ, Malpartida F, Hopwood DA (1985) The Streptomyces plasmid SCP2*: its functional analysis and development into useful cloning vectors. Gene 35:223–235
Messing J, Vieira J (1982) A new pair of M13 vectors for selecting either DNA strand of double-digest restriction fragments. Gene 19:269–276
Miller JH (1972) Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY
Mullen CA, Kilstrup M, Blaese RM (1992) Transfer of the bacterial gene for cytosine deaminase to mammalian cells confers lethal sensitivity to 5-fluorocytosine: a negative selection system. Proc Natl Acad Sci U S A 89:33–37
Nordstroem K, Austin SJ (1989) Mechanisms that contribute to the stable segregation of plasmids. Annu Rev Genet 23:37–69
O’Donovan GA, Neuhard J (1970) Pyrimidine metabolism in microorganisms. Bacteriol Rev 34:278–343
Rajeev L, Malanowska K, Gardner JF (2009) Challenging a paradigm: the role of DNA homology in tyrosine recombinase reactions. Microbiol Mol Biol Rev 73:300–309
Sambrook J, Russell DWDW (2001) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
Sandhu U, Cebula M, Behme S, Riemer P, Wordarczyk C, Metzger D, Reimann J, Schirmbeck R, Hauser H, Wirth D (2011) Strict control of transgene expression in a mouse model for sensitive biological applications based on RMCE compatible ES cells. Nucleic Acids Res 39:1
Sauer B (1992) Identification of cryptic lox sites in the yeast genome by selection for Cre-mediated chromosome translocations that confer multiple drug resistance. J Mol Biol 223:911–928
Schlake T, Bode J (1994) Use of mutated FLP recognition target (FRT) sites for the exchange of expression cassettes at defined chromosomal loci. Biochemistry 33:12746–12751
Schrempf H, Bujard H, Hopwood DA, Goebel W (1975) Isolation of covalently closed circular deoxyribonucleic acid from Streptomyces coelicolor A3(2). J Bacteriol 121:416–421
Sheren J, Langer SJ, Leinwand LA (2007) A randomized library approach to identifying functional lox site domains for the Cre recombinase. Nucleic Acids Res 35:5464–5473
Sorrell DA, Robinson CJ, Smith J-A, Kolb AF (2010) Recombinase mediated cassette exchange into genomic targets using an adenovirus vector. Nucleic Acids Res 38:123
Stark WM, Boocock MR, Sherratt DJ (1992) Catalysis by site-specific recombinases. Trends Genet 8:432–439
Strecker HJ (1957) The interconversion of glutamic acid and proline. I. The formation of Δ1-pyrroline-5-carboxylic acid from glutamic acid in Escherichia coli. J Biol Chem 225:825–834
Subramanya HS, Arciszewska LK, Baker RA, Bird LE, Sherratt DJ, Wigley DB (1997) Crystal structure of the site-specific recombinase, XerD. EMBO J 16:5178–5187
Suzuki N, Nonaka H, Tsuge Y, Inui M, Yukawa H (2005) New multiple-deletion method for the Corynebacterium glutamicum genome, using a mutant lox sequence. Appl Environ Microbiol 71:8472–8480
Suzuki N, Inui M, Yukawa H (2007) Site-directed integration system using a combination of mutant lox sites for Corynebacterium glutamicum. Appl Microbiol Biotechnol 77:871–878
Turan S, Kuehle J, Schambach A, Baum C, Bode J (2010) Multiplexing RMCE: versatile extensions of the Flp-recombinase-mediated cassette-exchange technology. J Mol Biol 402:52–69
Turan S, Galla M, Ernst E, Qiao J, Voelkel C, Schiedlmeier B, Zehe C, Bode J (2011) Recombinase-mediated cassette exchange (RMCE): traditional concepts and current challenges. J Mol Biol 407:193–221
Vogel HJ, Davis BD (1952) Glutamic γ-semialdehyde and Δ1-pyrroline-5-carboxylic acid, intermediates in the biosynthesis of proline. J Am Chem Soc 74:109–112
Warth L, Haug I, Altenbuchner J (2011) Characterization of the tyrosine recombinase MrpA encoded by the Streptomyces coelicolor A3(2) plasmid SCP2*. Arch Microbiol 193:187–200
Watson AT, Garcia V, Bone N, Carr AM, Armstrong J (2008) Gene tagging and gene replacement using recombinase-mediated cassette exchange in Schizosaccharomyces pombe. Gene 407:63–74
Yanisch-Perron C, Vieira J, Messing J (1985) Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103–119
Acknowledgments
The present work has been completed as part of a PhD thesis at Stuttgart University, Institute of Industrial Genetics (IIG). We would like to thank Prof. Dr. Ralf Mattes for his great and generous support during the past years of research.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Jean-Luc Pernodet.
Rights and permissions
About this article
Cite this article
Warth, L., Altenbuchner, J. The tyrosine recombinase MrpA and its target sequence: a mutational analysis of the recombination site mrpS resulting in a new left element/right element (LE/RE) deletion system. Arch Microbiol 195, 617–636 (2013). https://doi.org/10.1007/s00203-013-0910-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-013-0910-x