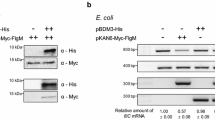
Abstract
In Legionella pneumophila, the regulation of the flagellum and the expression of virulence traits are linked. FleQ, RpoN and FliA are the major regulators of the flagellar regulon. We demonstrated here that all three regulatory proteins mentioned (FleQ, RpoN and FliA) are necessary for full in vivo fitness of L. pneumophila strains Corby and Paris. In this study, we clarified the role of FleQ for fliA expression from the level of mRNA toward protein translation. FleQ enhanced fliA expression, but FleQ and RpoN were not necessary for basal expression. In addition, we identified the initiation site of fliA in L. pneumophila and found a putative σ70 promoter element localized upstream. The initiation site was not influenced in the ΔfleQ or ΔrpoN mutant strain. We demonstrated that there is no significant difference in the regulation of fliA between strains Corby and Paris, but the FleQ-dependent induction of fliA transcription in the exponential phase is stronger in strain Paris than in strain Corby. In addition, we showed for the first time the presence of a straight hook at the pole of the non-flagellated ΔfliA and ΔfliD mutant strains by electron microscopy, indicating the presence of an intact basal body in these strains.





Similar content being viewed by others
References
Albert-Weissenberger C, Sahr T, Sismeiro O, Hacker J, Heuner K, Buchrieser C (2010) Control of flagellar gene regulation in Legionella pneumophila and its relation to growth phase. J Bacteriol 192(2):446–455
Bachman MA, Swanson MS (2001) RpoS co-operates with other factors to induce Legionella pneumophila virulence in the stationary phase. Mol Microbiol 40(5):1201–1214
Bachman MA, Swanson MS (2004) Genetic evidence that Legionella pneumophila RpoS modulates expression of the transmission phenotype in both the exponential phase and the stationary phase. Infect Immun 72(5):2468–2476
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Brüggemann H, Hagman A, Jules M, Sismeiro O, Dillies MA, Gouyette C, Kunst F, Steinert M, Heuner K, Coppee JY, Buchrieser C (2006) Virulence strategies for infecting phagocytes deduced from the in vivo transcriptional program of Legionella pneumophila. Cell Microbiol 8(8):1228–1240
Byrne B, Swanson MS (1998) Expression of Legionella pneumophila virulence traits in response to growth conditions. Infect Immun 66(7):3029–3034
Cazalet C, Rusniok C, Brüggemann H, Zidane N, Magnier A, Ma L, Tichit M, Jarraud S, Bouchier C, Vandenesch F, Kunst F, Etienne J, Glaser P, Buchrieser C (2004) Evidence in the Legionella pneumophila genome for exploitation of host cell functions and high genome plasticity. Nat Genet 36(11):1165–1173
Dalebroux ZD, Edwards RL, Swanson MS (2009) SpoT governs Legionella pneumophila differentiation in host macrophages. Mol Microbiol 71(3):640–658
Dalebroux ZD, Svensson SL, Gaynor EC, Swanson MS (2010a) ppGpp conjures bacterial virulence. Microbiol Mol Biol Rev 74(2):171–199
Dalebroux ZD, Yagi BF, Sahr T, Buchrieser C, Swanson MS (2010b) Distinct roles of ppGpp and DksA in Legionella pneumophila differentiation. Mol Microbiol 76(1):200–219
Dietrich C (2000) Molekularbiologische Studien zur Bedeutung der Flagelle fuer die Virulenz von Legionella pneumophila. University of Wuerzburg, Wuerzburg
Dietrich C, Heuner K, Brand BC, Hacker J, Steinert M (2001) Flagellum of Legionella pneumophila positively affects the early phase of infection of eukaryotic host cells. Infect Immun 69(4):2116–2122
Edelstein PH (1981) Improved semiselective medium for isolation of Legionella pneumophila from contaminated clinical and environmental specimens. J Clin Microbiol 14(3):298–303
Edwards RL, Dalebroux ZD, Swanson MS (2009) Legionella pneumophila couples fatty acid flux to microbial differentiation and virulence. Mol Microbiol 71(5):1190–1204
Eylert E, Herrmann V, Jules M, Gillmaier N, Lautner M, Buchrieser C, Eisenreich W, Heuner K (2010) Isotopologue profiling of Legionella pneumophila: role of serine and glucose as carbon substrates. J Biol Chem 285(29):22232–22243
Gal-Mor O, Segal G (2003) The Legionella pneumophila GacA homolog (LetA) is involved in the regulation of icm virulence genes and is required for intracellular multiplication in Acanthamoeba castellanii. Microb Pathog 34(4):187–194
Glöckner G, Albert-Weissenberger C, Weinmann E, Jacobi S, Schunder E, Steinert M, Hacker J, Heuner K (2008) Identification and characterization of a new conjugation/type IVA secretion system (trb/tra) of Legionella pneumophila Corby localized on two mobile genomic islands. Int J Med Microbiol 298(5–6):411–428
Hales LM, Shuman HA (1999) The Legionella pneumophila rpoS gene is required for growth within Acanthamoeba castellanii. J Bacteriol 181(16):4879–4889
Hall J, Voelz H (1985) Bacterial endosymbionts of Acanthamoeba sp. J Parasitol 71(1):89–95
Hammer BK, Swanson MS (1999) Co-ordination of Legionella pneumophila virulence with entry into stationary phase by ppGpp. Mol Microbiol 33(4):721–731
Hammer BK, Tateda ES, Swanson MS (2002) A two-component regulator induces the transmission phenotype of stationary-phase Legionella pneumophila. Mol Microbiol 44(1):107–118
Hanahan D (1983) Studies on transformation of Escherichia coli with plasmids. J Mol Biol 166(4):557–580
Helbig JH, Benson RF, Pelaz C, Jacobs E, Lück PC (2007) Identification and serotyping of atypical Legionella pneumophila strains isolated from human and environmental sources. J Appl Microbiol 102(1):100–105
Heuner K, Albert-Weissenberger C (2008) The flagellar regulon of Legionella pneumophila and the expression of virulence traits. In: Heuner K, Swanson M (eds) Legionella—Molecular Microbiology. Horizon Scientific Press, UK, pp 101–121
Heuner K, Bender-Beck L, Brand BC, Luck PC, Mann KH, Marre R, Ott M, Hacker J (1995) Cloning and genetic characterization of the flagellum subunit gene (flaA) of Legionella pneumophila serogroup 1. Infect Immun 63(7):2499–2507
Heuner K, Hacker J, Brand BC (1997) The alternative sigma factor sigma28 of Legionella pneumophila restores flagellation and motility to an Escherichia coli fliA mutant. J Bacteriol 179(1):17–23
Heuner K, Meltzer U, Choi BK, Gobel UB (2001) Outer sheath associated proteins of the oral spirochete Treponema maltophilum. FEMS Microbiol Lett 197(2):187–193
Heuner K, Dietrich C, Skriwan C, Steinert M, Hacker J (2002) Influence of the alternative sigma(28) factor on virulence and flagellum expression of Legionella pneumophila. Infect Immun 70(3):1604–1608
Hirano T, Yamaguchi S, Oosawa K, Aizawa S (1994) Roles of FliK and FlhB in determination of flagellar hook length in Salmonella typhimurium. J Bacteriol 176(17):5439–5449
Horwitz MA (1983) Formation of a novel phagosome by the Legionnaires’ disease bacterium (Legionella pneumophila) in human monocytes. J Exp Med 158(4):1319–1331
Horwitz MA, Maxfield FR (1984) Legionella pneumophila inhibits acidification of its phagosome in human monocytes. J Cell Biol 99(6):1936–1943
Horwitz MA, Silverstein SC (1980) Legionnaires’ disease bacterium (Legionella pneumophila) multiples intracellularly in human monocytes. J Clin Investig 66(3):441–450
Jacobi S, Schade R, Heuner K (2004) Characterization of the alternative sigma factor sigma54 and the transcriptional regulator FleQ of Legionella pneumophila, which are both involved in the regulation cascade of flagellar gene expression. J Bacteriol 186(9):2540–2547
Jepras RI, Fitzgeorge RB, Baskerville A (1985) A comparison of virulence of two strains of Legionella pneumophila based on experimental aerosol infection of guinea-pigs. J Hyg 95(1):29–38
Kagawa H, Aizawa SI, Asakura S (1979) Transformations in isolated polyhooks. J Mol Biol 129(2):333–336
Lacour S, Landini P (2004) SigmaS-dependent gene expression at the onset of stationary phase in Escherichia coli: function of sigmaS-dependent genes and identification of their promoter sequences. J Bacteriol 186(21):7186–7195
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227(5259):680–685
McCarter LL (2006) Regulation of flagella. Curr Opin Microbiol 9(2):180–186
Molofsky AB, Swanson MS (2003) Legionella pneumophila CsrA is a pivotal repressor of transmission traits and activator of replication. Mol Microbiol 50(2):445–461
Molofsky AB, Swanson MS (2004) Differentiate to thrive: lessons from the Legionella pneumophila life cycle. Mol Microbiol 53(1):29–40
Molofsky AB, Shetron-Rama LM, Swanson MS (2005) Components of the Legionella pneumophila flagellar regulon contribute to multiple virulence traits, including lysosome avoidance and macrophage death. Infect Immun 73(9):5720–5734
Nyström T (2004) Growth versus maintenance: a trade-off dictated by RNA polymerase availability and sigma factor competition? Mol Microbiol 54(4):855–862
Potvin E, Sanschagrin F, Levesque RC (2008) Sigma factors in Pseudomonas aeruginosa. FEMS Microbiol Rev 32(1):38–55
Rasis M, Segal G (2009) The LetA-RsmYZ-CsrA regulatory cascade, together with RpoS and PmrA, post-transcriptionally regulates stationary phase activation of Legionella pneumophila Icm/Dot effectors. Mol Microbiol 72(4):995–1010
Rowbotham TJ (1986) Current views on the relationships between amoebae, legionellae and man. Isr J Med Sci 22(9):678–689
Sahr T, Brüggemann H, Jules M, Lomma M, Albert-Weissenberger C, Cazalet C, Buchrieser C (2009) Two small ncRNAs jointly govern virulence and transmission in Legionella pneumophila. Mol Microbiol 72(3):741–762
Sambrook J, Russell D (2000) Molecular cloning: a laboratory manual, 3 Vol. Cold Spring Harbor Laboratory, NY
Sauer JD, Bachman MA, Swanson MS (2005a) The phagosomal transporter A couples threonine acquisition to differentiation and replication of Legionella pneumophila in macrophages. Proc Nat Acad Sci USA 102(28):9924–9929
Sauer JD, Shannon JG, Howe D, Hayes SF, Swanson MS, Heinzen RA (2005b) Specificity of Legionella pneumophila and Coxiella burnetii vacuoles and versatility of Legionella pneumophila revealed by coinfection. Infect Immun 73(8):4494–4504
Schunder E, Adam P, Higa F, Remer KA, Lorenz U, Bender J, Schulz T, Flieger A, Steinert M, Heuner K (2010) Phospholipase PlaB is a new virulence factor of Legionella pneumophila. Int J Med Microbiol 300(5):313–323
Shimizu R, Taguchi F, Marutani M, Mukaihara T, Inagaki Y, Toyoda K, Shiraishi T, Ichinose Y (2003) The DeltafliD mutant of Pseudomonas syringae pv. tabaci, which secretes flagellin monomers, induces a strong hypersensitive reaction (HR) in non-host tomato cells. Mol Genet Genomics 269(1):21–30
Stone BJ, Kwaik YA (1999) Natural competence for DNA transformation by Legionella pneumophila and its association with expression of type IV pili. J Bacteriol 181(5):1395–1402
Sturgill-Koszycki S, Swanson MS (2000) Legionella pneumophila replication vacuoles mature into acidic, endocytic organelles. J Exp Med 192(9):1261–1272
Tiaden A, Spirig T, Weber SS, Bruggemann H, Bosshard R, Buchrieser C, Hilbi H (2007) The Legionella pneumophila response regulator LqsR promotes host cell interactions as an element of the virulence regulatory network controlled by RpoS and LetA. Cell Microbiol 9(12):2903–2920
Zusman T, Gal-Mor O, Segal G (2002) Characterization of a Legionella pneumophila relA insertion mutant and roles of RelA and RpoS in virulence gene expression. J Bacteriol 184(1):67–75
Acknowledgments
We would like to thank Ursula Erikli for careful reading of the manuscript. This work received financial support from the Robert Koch-Institute and by grant HE2854/5-1 + 2 from the Deutsche Forschungsgemeinschaft DFG (Bonn, Germany) to K.H.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by S. Suerbaum.
Electronic supplementary material
Below is the link to the electronic supplementary material.
203_2012_833_MOESM1_ESM.pdf
Online Resource 1 Reverse transcription (RT-)PCR analysis of L. pneumophila Corby. Bacteria were grown in AYE medium and samples were taken during different growth phases and prepared for RT-PCR as well as Western blot analysis. For RT-PCR experiments, whole-cell RNA was isolated and equal amounts (10 ng) were used for amplification. For all samples, RT-PCR was performed at (a) 24 cycles, (b) 27 cycles or (c) 30 cycles, respectively. Results were carried out in at least two independent experiments. Abbreviations: E, exponential growth phase; LE, late exponential growth phase; PE, post-exponential growth phase; S, stationary growth phase (PDF 285 kb)
203_2012_833_MOESM2_ESM.pdf
Online Resource 2 Determination of the transcriptional start site of fliA via 5′RACE experiments – raw data. RNA from late exponentially grown L. pneumophila Corby wild type (LpC wt; a, b), LpC ΔfleQ (a, c) and LpC ΔrpoN (a, d) mutant strains was transcribed in cDNA, polyA-tailed and amplified. (a) All RNA samples contained a control RNA to verify the 5′RACE reaction via PCR. cDNA synthesis (K1; primers: neo2/rev and neo3/for; product: 157 bp), cDNA purification (K2; primers: neo2/rev and neo3/for; product: 157 bp) and d(A)-tailing reaction (K3; primers: neo2/rev and oligo d(T)-anchor; product: 293 bp) were verified with primers specific to the control RNA, respectively. The PCR product (R; primers: oligo d(T)-anchor and FliA_RACE_5R; product: ~ 500 bp) was sequenced using the anti-sense primer FliA_RACE_6R. The identified transcriptional start site is indicated (+1; b, c, d) (PDF 278 kb)
Rights and permissions
About this article
Cite this article
Schulz, T., Rydzewski, K., Schunder, E. et al. FliA expression analysis and influence of the regulatory proteins RpoN, FleQ and FliA on virulence and in vivo fitness in Legionella pneumophila . Arch Microbiol 194, 977–989 (2012). https://doi.org/10.1007/s00203-012-0833-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-012-0833-y