Abstract
Background
Vibrio cholerae, a Gram-negative bacterium, is highly motile owing to the presence of a single polar flagellum. The global anaerobiosis response regulator, ArcA regulates the expression of virulence factors and enhance biofilm formation in V. cholerae. However, the function of ArcA for the motility of V. cholerae is yet to be elucidated. CytR, which represses nucleoside uptake and catabolism, is known to play a chief role in V. cholerae pathogenesis and flagellar synthesis but the mechanism that CytR influences motility is unclear.
Results
In this study, we found that the ΔarcA mutant strain exhibited higher motility than the WT strain due to ArcA directly repressed flrA expression. We further discovered that CytR directly enhanced fliK expression, which explained why the ΔcytR mutant strain was retarded in motility. On the other hand, cytR was a direct ArcA target and cytR expression was directly repressed by ArcA. As expected, cytR expression was down-regulated.
Conclusions
Overall, ArcA plays a critical role in V. cholerae motility by regulating flrA expression directly and fliK indirectly in the manner of cytR.
Similar content being viewed by others

Background
V. cholerae is ubiquitous in aquatic environments and intestines of host [1]. The flagellum of V. cholerae is a complex self-assembling organelle that is attached to the cell surface and allows bacterial cells to move in their environment [2]; moreover, it plays a pivotal role in substrates adhesion, biofilm formation, and virulence [3,4,5,6]. Elucidating the mechanisms underlying the regulation of flagellum should enhance our understanding of the lifecycle of V. cholerae both in the intestinal and aquatic phases. V. cholerae flagellar genes are expressed within a fourtiered transcriptional hierarchy [4]. The sole Class I gene encodes the σ54-dependent transcriptional activator FlrA, which is the master regulator of the flagellar hierarchy because without it no flagellar genes are expressed [7]. FliK is in the Class III level of the flagellar hierarchy. The assembly of a flagellum occurs in a number of stages, and FliK is the ‘‘checkpoint control’’ protein When the flagellar hook substructure has reached its optimal length, FliK then terminates hook export and assembly and transmits a signal to begin filament export, in the final stage of the flagellar biosynthesis [8].
The ArcA/ArcB two-component system evidently has a key role in the response to oxygen. The membrane sensor protein ArcB is phosphorylated, resulting in the transfer of its phosphoryl group to ArcA. Phosphorylated ArcA is subsequently activated as a transcription factor, resulting in the up-or downregulation of several downstream genes [9, 10]. In Escherichia coli, approximately 50% genes whose expression levels are affected due to aerobic to anaerobic transitions are also affected by ArcA; in total 1139 genes in the E. coli genome are in fact either directly or indirectly regulated by ArcA [11]. In Salmonella Typhimurium, ArcA has been shown to affect metabolic processes, stress response, and surface adherence [12]. A study reported that the ΔarcA mutant strain lacked flagella in S. enterica sv. Typhimurium and was thus non-motile [13]. In E. coli, ArcA enhances motility by increasing fliA expression [14]. Further ArcA was found to directly repress the expression of motA, motB, and cheA in avian pathogenic E. coli, but it did not affect the expression of flagella genes [15]. Considering that the function of ArcA in V. cholerae motility remains unexplored, in this study, we aimed to determine the relationship between ArcA and V. cholerae motility.
The CytR repressor belongs to the LacI family, and possesses, like the other members, an N-terminal helix–turn–helix (HTH) DNA-binding motif [16]. However, unlike a typical bacterial repressor, the CytR repressor and the cAMP receptor protein (CRP) bind cooperatively to several promoters in E. coli to repress transcription initiation [17]. In addition, researches had shown that the CRP protein and the CytR regulator can act either to repress or to activate transcription depending on the context [18]. CytR negatively regulates the genes that are involved in nucleosides uptake and catabolism [19]. In uropathogenic E. coli, CytR is a modulator of flagellar expression activated by CRP. The ΔcytR mutant strain was observed to show higher motility and flagellar expression [20]; further, CytR bound to the upstream region of flhD, which encodes the master regulator for flagellar expression. In V. cholerae, CytR positively regulates competence genes, type VI secretion operons, and chitinases [21]. Moreover, it plays an important role in V. cholerae pathogenesis and flagella synthesis [22]. In V. cholrae, the ΔcytR mutant strain was found to show downregulated expression levels of the class II flagellar genes flrB and flrC, and several class III flagellar genes [22]. Nevertheless, further studies are warranted to comprehensively understand this process. In the current study, we identified that low oxygen levels enhanced arcA expression, but repressed cytR expression. Furthermore, ArcA reduces V. cholerae motility not only in the manner of directly regulating flrA, but also in the manner of indirectly regulating fliK via directly binding and regulating cytR. Collectively, our findings enhance our understanding of how ArcA and CytR collaboratively modulate V. cholerae motility.
Results
ArcA repressed V. cholerae motility
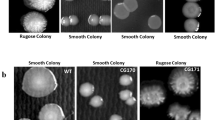
In E. coli and S. enterica sv. Typhimurium, ArcA acts as the positive regulator of motility [13, 14]. To evaluate the role of ArcA in V. cholerae motility, we investigated the surface motility of ΔarcA mutant strain on soft agar plates in aerobic conditions. The motility zones of the ΔarcA mutant strain (diameter, 2.25 ± 0.15 cm) were larger than those of the WT strain (diameter, 1.27 ± 0.17 cm) (Figs. 1a and 1b). Moreover, we constructed the arcA complemented strain ΔarcA::ParcA containing a functional copy of the arcA sequence, using the plasmid pBAD33; and found the complementation strain (diameter, 1.51 ± 0.11 cm) restore the motility to the WT level (p = 0.0913) (Figs. 1a and 1b). These data indicated that ArcA functions as a negative regulator of V. cholerae motility.
ArcA directly repressed flrA expression
The increase in the motility of the ΔarcA mutant strain could be attributed to lack of flagella (fla phenotype), loss of motility (mot phenotype), or loss of chemotaxis (che phenotype) [14, 23]. As previous reports, ArcA is proposed to bind to a conservative sequence (5’-TGTTA-3’) in E. coli [24]. Then, A search for the conservative ArcA binding sites in E. coli at flrA promoter region was performed using Virtual Footprint 3.0. We found a similar sequence (TGTTC-AAACGGTGCAACCACAACT-TCTTA) with a 19 base-spacing at positions -14 to -42 on the upstream region of flrA that ArcA probably binds (Fig. S10). To determine the precise cause, the EMSA screening was performed to assess whether ArcA directly binds to the promoter region of the 16 different core regulons in the flagellar heirarchy, and found that the phosphorylated ArcA directly bound to the promoter region of flrA (Fig. 2a), not with the other 15 ones. The following qRT-PCR indicated that flrA expression was increased by approximately 3.4-fold in the ΔarcA mutant strain compared with that in the WT strain in aerobic conditions (Fig. 2b). Moreover, we constructed the arcA complemented strain ΔarcA::ParcA containing a functional copy of the arcA gene, using the plasmid pBAD33; and found flrA expression in the complementation strain was restored to the WT level (p = 0.1320) (Fig. 2b). Furthermore, the promoter-lux fusion reporter plasmid was constructed with the promoter of flrA inserted into pMS402, and found that the activity of flrA promoter-lux was approximately up-regulation by 1.5-fold in the ΔarcA mutant strain compared to that in the WT (Fig. 2c). These data indicated that ArcA functions as a negative regulator of V. cholerae motility by directly repressing the expression of flrA.
ArcA directly represses flrA expression. a. The EMSA between phosphorylated ArcA protein and the flrA promoter. The concentration of phosphorylated ArcA protein increased gradually (0 to 2.4 μM), and the amount of promoter DNA used in each reaction was 50 ng. 4.5S RNA served as the negative control. b. mRNA levels of flrA in the WT,,ΔarcA mutant strain and arcA complementary strain ΔarcA::ParcA in aerobic conditions. *, p < 0.05; **, p < 0.01; ***, p < 0.001; NS, no significance, p > 0.05. c. Expression of flrA-lux in WT strain and ΔarcA mutant strain. CPS (counts per second) values represent relative promoter-lux activities. All experiments were independently repeated at least three times. Values represent means ± SEM
CytR enhanced the fliK expression and activated by CRP
Previous studies reported that the ΔcytR mutant strain shows reduced motility and the expression of flagellar-synthesis regulatory genes flrBC and class III flagellar-synthesis genes were reduced in the ΔcytR mutant strain [22]. We hypothesized that CytR modulates flagellar regulatory genes by directly binding to their promoter regions. CytR is proposed to bind to octameric inverted or direct repeats containing the consensus half sites (5’-TGCAA-3’) with variable spacing in E. coli [16]. We found a similar sequence (TGCAA-TAAAACCTTCACTTGGCTTACTTC-TTGCT) with a 24 base-spacing at positions -15 to -48 on the upstream region of fliK that CytR probably binds (Fig S11). This site overlays 18 bases of CRP-binding site (TGGAT-GCAATAAAACCT-TCACT) located at position -31 to -52 (Fig. S2) [24].
The EMSA was performed to assess whether CytR directly binds to the promoter region of the 16 different core regulons in the flagellar heirarchy, and found that the CytR protein directly bound to the promoter region of fliK (Fig. 3a), not with the other 15 ones. Studies of gene regulation have revealed that the CRP protein and the CytR regulator can act either to repress or to activate transcription depending on the context [18]. When CytR binds to the promoter region of target genes, it forms a complex with CRP and RNA polymerase [25]. To determine if the transcriptional regulation of fliK by CytR occurs in the same manner as that of the above cases, we also observed CRP protein only (Fig. 3b) and co-binding of the CytR and CRP proteins by EMSA assay (Fig. 3c).
CytR directly promotes fliK expression. a. The EMSA of CytR with the fliK promoter. The concentration of CytR protein increased gradually (0 to 3.6 μM), and the amount of promoter DNA used in each reaction was 50 ng. b. The EMSA of CRP with the fliK promoter. The concentration of CRP protein increased gradually (0 to 4 μM), and the amount of promoter DNA used in each reaction was 50 ng. The concentration of cAMP used in each reaction was 0.1 mM. c. The EMSA of CytR and CRP with the fliK promoter. The concentration of CytR and CRP protein increased gradually (0 to 3.6 or 4 μM), and the amount of promoter DNA used in each reaction was 50 ng. The concentration of cAMP used in each reaction was 0.1 mM. d. mRNA levels of fliK in the WT, ΔcytR mutant strain and the complementary strain ΔcytR::PcytR. *, p < 0.05; **, p < 0.01; ***, p < 0.001; NS, no significance, p > 0.05. e. Expression of fliK-lux in WT strain and ΔcytR mutant strain. CPS (counts per second) values represent relative promoter-lux activities. All experiments were independently repeated at least three times. Values represent means ± SEM
qRT-PCR revealed that fliK expression was approximately down-regulated by 2.4-fold in the ΔcytR mutant strain compared with that in the WT strain in aerobic conditions (Fig. 3d). Moreover, we constructed the cytR complemented strain ΔcytR::PcytR containing a functional copy of the cytR gene, using the plasmid pBAD33; and found fliK expression in the complementation strain was restored to the WT level (p = 0.1126) (Fig. 3d). Furthermore, the promoter-lux fusion reporter plasmid was constructed with the promoter of fliK inserted into pMS402 and found that the activity of fliK promoter-lux was approximately down-regulation by 1.5-fold in the ΔcytR mutant strain compared to that in the WT (Fig. 3e). As a whole, CytR and CRP cooperatively bind to the upstream region of fliK, then CytR enhance fliK expression in the presence of CRP as a modulator.
ArcA directly repressed cytR expression
In addition to ArcA and CytR being related to the V. cholerae motility, we also found that CytR is a new downstream regulatory gene of ArcA. As previous reports, ArcA is proposed to bind to a conservative sequence (5’-TGTTA-3’) in E. coli [24]. Then, A search for the conservative ArcA binding sites in E. coli at cytR promoter region was performed using Virtual Footprint 3.0. We found a similar sequence (TGTTA-ATTTTGTCAGCAAATTAATGC-TTATTA) with a 21 base-spacing at positions -11 to -42 on the upstream region of cytR that ArcA probably binds (Fig. S12). Then EMSA showed that the phosphorylated ArcA directly binds to the promoter of cytR (Fig. 4a). qRT-PCR was was performed in both aerobic and anaerobic conditions. The results showed that the cytR expression was increased by 2.1-fold in aerobic conditions, and 5.0-fold in anaerobic condition in the ΔarcA mutant strain compared to that in the WT strain (Fig. 4b and 4c). Moreover, we constructed the arcA complemented strain ΔarcA::ParcA containing a functional copy of the arcA sequence, using the plasmid pBAD33; and found cytR expression in the complementation strain restore to the WT level in aerobic conditions (p = 0.6134) and anaerobic conditions(p = 0.3347) (Fig. 4b and 4c). Furthermore, the promoter-lux fusion reporter plasmid was constructed with the promoter of cytR inserted into pMS402 and found that the activity of cytR promoter-lux was approximately up-regulation by 1.3-fold in the ΔarcA mutant strain compared to that in the WT (Fig. 4d). These data indicated that ArcA repressed cytR expression by directly binding to its promoter region.
ArcA directly represses cytR expression. a. The EMSA between phosphorylated ArcA protein and the cytR promoter. The concentration of phosphorylated ArcA protein increased gradually (0 to 6 μM), and the amount of promoter DNA used in each reaction was 50 ng. b. mRNA levels of cytR in the WT, ΔarcA mutant strain and the complementary strain ΔarcA::ParcA in aerobic condition. *, p < 0.05; **, p < 0.01; ***, p < 0.001; NS, no significance, p > 0.05. c. mRNA levels of cytR in the WT, ΔarcA mutant strain and the complementary strain ΔarcA::ParcA in anaerobic condition. *, p < 0.05; **, p < 0.01; ***, p < 0.001; NS, no significance, p > 0.05. d. Expression of cytR-lux in WT strain and ΔarcA mutant strain. CPS (counts per second) values represent relative promoter-lux activities. All experiments were independently repeated at least three times, and the data shown represent comparable results. Values represent means ± SEM
Discussion
ArcA and CytR involved in multiple regulation in V. cholerae. ArcA, being a regulator, its function in V. cholerae motility is still not known. Here, we for the first time aimed to understand the role of this multifunctional transcription factor in V. cholerae motility. Our results aslo suggest an expanded role of ArcA in V. cholerae in the manner of cytR and further evidence the relationship between CytR and V. cholerae motility.
More than 50 genes are involved in flagella synthesis and regulation in V. cholerae [7]. The flagella-synthesis genes in V. cholerae are categorized into a four transcriptional hierarchy [26]. The σ54-dependent transcriptional activator FlrA is the only class I gene in this hierarchy [27]. FlrA is the master regulator of the V. cholerae flagellar transcription hierarchy because it is important for the expression of all other flagellar genes. The fliK operon is transcribed from a class III promoter [28]. Flagellar assembly occurs in a number of stages, and in this process, the ‘‘checkpoint control’’ protein FliK functions in detecting when the flagellar hook substructure has reached its optimal length. FliK then terminates hook export and assembly and transmits a signal to begin filament export [8, 29].
In E. coli, the ΔarcA mutant strain has been reported to show loss of motility, with ArcA being necessary for the expression of fliA, but not for that of the master regulators flhDC [30]. In avian pathogenic E. coli, ArcA directly regulates the expression of motA, motB, and cheA [15], whereas in S. enterica sv. Typhimurium, it activates class II and III flagellar genes and seems to slightly repress flhDC [31]. In contrast, in V. cholerae, we found that ArcA reduced motility by a directly way of repressing the expression of the class I flagellar regulatory gene flrA, and an indirrectly way of repressing the expression of the class III flagellar regulatory gene fliK via cytR (Fig. 5). The expression of cytR was up-regulated by 2.1-fold in ΔarcA stain compared to the WT under the aerobic condition (Fig. 4b); likewise, up-regulated by 5.0-fold under the anaerobic condition (Fig. 4c). So the further qRT-PCR was performed on the cytR in ΔarcA stain under both the aerobic and anaerobic conditions, and the data showed that the expression of cytR between these two conditions was not significantly different. In other word, the repression of ArcA on cytR was consistent, no matter under the aerobic or anaerobic conditions.
In uropathogenic E. coli, CytR evidently represses motility and flagellar expression by directly binding to the upstream region of flhD, which encodes the master regulator for flagellar expression [20]. In the ΔcytR mutant strain of V. cholerae, qRT-PCR showed that the expression of the class II flagellar genes flrB and flrC, and that of several class III flagellar genes was downregulated [32]. We further investigated the target binding site of CytR and found that CytR bound to the promoter region of fliK and acts as a positive regulator of fliK (Fig. 5).
Our results suggest an extended role of ArcA in V. cholerae motility, and found its new downstream regulatory gene, cytR, which is also a global regulator and influences the motility by directly increasing flik expression.
Conclusions
In this work, we report that the global regulators ArcA and CytR collaboratively modulate V. cholerae motility. Here, we provide evidence that ArcA plays a fundamental role in V. cholerae motility by regulating the expression of flrA directly and fliK indirectly in the manner of cytR.
Materials and methods
Bacterial strains, plasmids, and growth conditions
All strains and plasmids used in this study are shown in Table 1. In aerobic condition, all strains were grown overnight at 37 °C in lysogeny broth/agar. In anaerobic condition, bacterial cultures in an anaerobic incubator were grown in the presence of 1 g/l cysteine and 1 mg/l resazurin. Antibiotics were added, as required 40 μg/ml, polymyxin B or 25 μg/ml, chloramphenicol. All chemicals were purchased from Sigma (St. Louis, MO, USA).
Construction of the deletion mutant of ArcA and its complementation
The ArcA isogenic deletion mutant was constructed using the suicide plasmid pRE112 method, as previously described [33]. Briefly, 1) the recombinant plasmid pRE112-ΔarcA-V.cholerae was constructed and transformed into E. coli λpir; 2) intergeneric conjugation between E. coli and V. cholerae. DNA sequencing was performed to confirm the sequences of the constructed deletion plasmids. The ΔarcA mutant strain were complemented with the plasmid cloned into the vector pBAD33. The complemented strain was constructed using a previously reported procedure [34]. Table 2 lists the primers used in this study.
Soft agar motility assay
V. cholerae strains were grown in LB broth for overnight and inoculated (1 μl) into freshly poured 0.3% agar plates, followed by incubation and grown at 30 °C for 24 h. The diameters of motility zones at least six independent colonies were averaged [35, 36].
RNA isolation and quantitative real time PCR (qRT-PCR)
Bacterial cultures were grown in LB medium aerobically or anaerobically at 37 °C to the mid-logarithmic phase (OD600 approximately 0.6). Total RNA was extracted using TRIzol (Invitrogen, Waltham, MA, USA, #15,596–018), as per manufacturer instructions. cDNA was synthesized using a Prime Script RT Reagent Kit with gDNA Eraser (Takara, Shiga, Japan). qRT-PCR was performed on an Applied Biosystems 7500 sequence detection system with SYBR green fluorescence dye. The 16 s rRNA gene was used as the reference control for sample normalization [37]. Table 2 lists the primers used in this study. The relative expression levels of target transcripts were calculated according to the 2−∆∆CT method [38]. Each experiment was performed in triplicate. Expression changes of > twofold with p < 0.05 were considered statistically significant.
Electrophoretic mobility shift assay (EMSA)
A sequence encoding a ArcA/CytR/CRP-His6 fusion protein was cloned into vector pET-28a, expressed in E. coli BL21 (DE3), and purified using an Ni–NTA-Sefinose Column in accordance with the protocol provided by the manufacturer [37, 39]. EMSA was performed by adding increasing amounts of purified phosphorylated ArcA protein (0, 1.5, 3.0, 4.5, and 6.0 µM or 0, 0.6, 1.2, 1.8 and 2.4 µM) to cytR or flrA DNA fragments (50 ng) in a binding buffer [10 mM Tris–HCl (pH 8.0), 1 mM EDTA, 1 mM DTT, 50 mM KCl, 50 μg/mL BSA, 10% glycerol] supplemented with 20 nM acetyl phosphate [39], followed by incubation for 40 min at room temperature. Similarly, fliK DNA fragments (50 ng) were incubated with increasing amounts of 6 × His-tagged CytR or CRP protein (0, 1.2, 2.4, and 3.6 µM or 0, 2, 3, and 4 µM) in a binding buffer [20 mM Tris–HCl (pH 7.5), 50 mM KCl, 1 mM dithiothreitol, 200 µM cAMP, and 10% glycerol] [20], followed by incubation for 40 min at room temperature. The concentration of cAMP used in each reaction was 0.1 mM. 4.5 s RNA served as the negative control. The reaction mixtures were then electrophoresed on a 6% native polyacrylamide gel. Protein was visualized using a Typhoon phosphorimager (GE Healthcare, Chicago, IL, USA).
Bioluminescent reporter assays
The procedures of the lux bioluminescent reporter assay were described in previous study [40]. Briefly, bacterial cultures were grown in LB medium at 37 °C to the mid-logarithmic phase (OD600 approximately 0.6). The cultures were transferred into a black 96-well plate with a transparent bottom. Promoter activities were measured and bacterial growth was measured by OD600 in a Synergy 2 plate reader (BioTek) at the same time.
Statistical analyses
All data are expressed as means ± standard deviation (SD). Differences between two groups were evaluated using independent-samples t-test or Mann–Whitney U test. Values of p ≤ 0.05, 0.01, or 0.001 were considered to be statistically significant (*), highly significant (**), or extremely significant (***), respectively.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Abbreviations
- ArcA:
-
Anoxic redox control cognate response regulator
- CRP:
-
Cyclic-AMP receptor protein
- CytR:
-
Cytidine repressor
- WT:
-
Wild-type
- ΔarcA::
-
ArcA isogenic deletion mutant strain;
- ΔarcA::
-
ParcA, complementation strain of arcA
- LB:
-
Luria–Bertani
- EMSA:
-
Electrophoretic mobility shift
References
Weil AA, Becker RL, Harris JB. Vibrio cholerae at the intersection of immunity and the microbiome. mSphere. 2019;4(6):e00597 19.
Klose KE, Mekalanos JJ. Distinct roles of an alternative sigma factor during both free-swimming and colonizing phases of the Vibrio cholerae pathogenic cycle. Mol Microbiol. 1998;28(3):501–20.
Attridge SR, Rowley DR. The Role of the Flagellum in the Adherence of Vibrio cholerae. J Infect Dis. 1983;147(5):864–72.
Syed KA, Beyhan S, Correa N, Queen J, Liu J, Peng F, Satchell K, et al. The Vibrio cholerae flagellar regulatory hierarchy controls expression of virulence factors. J Bacteriol. 2009;191(21):6555–70.
Soutourina OA, Bertin PN. Regulation cascade of flagellar expression in Gram- negative bacteria. FEMS Microbiol Rev. 2003;27(4):505–23.
Guttenplan SB, Kearns DB. Regulation of flagellar motility during biofilm formation. FEMS Microbiol Rev. 2013;37(6):849–71.
Echazarreta MA, Klose KE. Vibrio Flagellar Synthesis. Front Cell Infect Microbiol. 2019;9:131.
Waters RC, O’Toole PW, Ryan KA. The FliK protein and flagellar hook-length control. Protein Sci. 2007;16(5):769–80.
Teran-Melo JL, Peña-Sandoval GR, Silva-Jimenez H, Rodriguez C, Alvarez AF, Georgellis D. Routes of phosphoryl-group transfer during signal transmission and signal decay in the dimeric sensor histidine kinase ArcB. J Biol Chem. 2018;29(34):13214–23.
Jeon Y, Lee YS, Han JS, Kim JB, Hwang DS. Multimerization of phosphorylated and non-phosphorylated ArcA is necessary for the response regulator function of the Arc two-component signal transduction system. J Biol Chem. 2001;276(44):40873–9.
Salmon KA, Hung SP, Steffen NR, Krupp R, Baldi P, Hatfield GW, et al. Global gene expression profiling in Escherichia coli K12: Effects of oxygen availability and ArcA. J Biol Chem. 2005;280(15):15084–96.
Wang Z, Sun J, Xia T, Liu Y, Fu J, Kei LY, et al. Proteomic delineation of the ArcA regulon in Salmonella Typhimurium during anaerobiosis. Mol Cell Proteomics. 2018;17(10):1937–47.
Steffen P, Andres VT, Fink RC, Evans MR, Jessica JC, Michael MC, et al. Analysis of the ArcA regulon in anaerobically grown Salmonella enterica sv. Typhimurium BMC Microbiol. 2011;11(1):58.
Kato Y, Sugiura M, Mizuno T, Aiba H. Effect of the arcA Mutation on the expression of flagella genes in Escherichia coli. Biosci Biotechnol Biochem. 2007;1:77–83.
Jiang F, An C, Bao Y, Zhao X, Jernigan RL, Lithio A, et al. ArcA controls metabolism, chemotaxis, and motility contributing to the pathogenicity of Avian Pathogenic Escherichia coli. Infect Immun. 2015;83(9):3545–54.
Sernova NV, Gelfand MS, Roop I. Comparative genomics of CytR, an unusual member of the LacI Family of transcription factors. PLoS ONE. 2012;7(9):e44194.
Pedersen H, Valentin-Hansen P. Protein-induced fit: the CRP activator protein changes sequence-specific DNA recognition by the CytR repressor, a highly flexible LacI member. EMBO J. 1997;16(8):2108–18.
Rasmussen PB, Holst B, Valentin-Hansen P. Dual-function regulators: the cAMP receptor protein and the CytR regulator can act either to activate transcription depending on the context. Proc Natl Acad Sci U S A. 1996;93(19):10151–5.
Valentin-Hansen P, Sogaard-Andersen L, Pedersen H. A flexible partnership: the CytR anti-activator and the cAMP-CRP activator protein, comrades in transcription control. Mol Microbiol. 2010;20(3):461–6.
Hirakawa H, Takita A, Kato M, Mizumoto H, Tomita H. Roles of CytR, an anti-activator of cyclic-AMP receptor protein (CRP) on flagellar expression and virulence in uropathogenic Escherichia coli. Biochem Biophys Res Commun. 2020;521(3):555–61.
Watve SS, Jacob T, Hammer BK, Tom C. CytR is a global positive regulator of competence, type VI secretion, and chitinases in Vibrio cholerae. PLoS ONE. 2015;10(9):e0138834.
Das S, Chourashi R, Mukherjee P, Gope A, Chatterjee NS. Multifunctional transcription factor CytR of Vibrio cholerae is important for pathogenesis. Microbiology. 2020;166(12):1136–48.
Boin MA, Austin MJ, Häse C. Chemotaxis in Vibrio cholerae. FEMS Microbiol Lett. 2004;239(1):1–8.
Manneh-Roussel J, Haycocks JRJ, Magan A, Perez-Soto N, Voelz K, Camilli A, et al. cAMP receptor protein controls Vibrio cholerae gene expression in response to host colonization. MBio. 2018;9(4):e00966-e1018.
Chahla M, Wooll J, Laue TM, Nguyen N, Senear DF. Role of protein-protein bridging interactions on cooperative assembly of DNA-bound CRP-CytR-CRP complex and regulation of the Escherichia coli CytR regulon. Biochemistry. 2003;42(13):3812–25.
Soutourina OA, Bertin PN. Regulation cascade of flagellar expression in Gram-negative bacteria. FEMS Microbiol Rev. 2003;27(4):505–23.
Millikan DS, Ruby EG. FlrA, a σ[sup 54]-dependent transcriptional activator in Vibrio fischeri, is required for motility and symbiotic light-organ colonization. J Bacteriol. 2003;185(12):3547–57.
Kinoshita M, Tanaka S, Inoue Y, Namba K, Aizawa SI, Minamino T. The flexible linker of the secreted FliK ruler is required for export switching of the flagellar protein export apparatus. Sci Rep. 2020;10(1):838.
Moriya N, Minamino T, Hughes KT, Macnab RM, Namba K. The type III flagellar export specificity switch is dependent on FliK ruler and a molecular clock. J Mol Biol. 2006;359(2):466–77.
Kato Y, Sugiura M, Mizuno T, Aiba H. Effect of the arcA mutation on the expression of flagella genes in Escherichia coli. Biosci Biotechnol Biochem. 2007;71(1):77–83.
Evans MR, Fink RC, Vazquez-Torres A, Porwollik S, Jones-Carson J, McClelland M, et al. Analysis of the ArcA regulon in anaerobically grown Salmonella enterica sv. Typhimurium BMC Microbiol. 2011;11:58.
Iuchi S, Lin EC. Mutational analysis of signal transduction by ArcB, a membrane sensor protein responsible for anaerobic repression of operons involved in the central aerobic pathways in Escherichia coli. J Bacteriol. 1992;174(12):3972–80.
Xu T, Cao H, Zhu W, Wang M, Du Y, Yin Z, et al. RNA-seq-based monitoring of gene expression changes of viable but non-culturable state of Vibrio cholerae induced by cold seawater. Environ Microbiol Rep. 2018;10(5):594–604.
Vettiger A, Basler M. Type VI secretion system substrates are transferred and reused among sister cells. Cell. 2016;167(1):99-110.e12.
Bartlett TM, Bratton BP, Duvshani A, Miguel A, Sheng Y, Martin NR, et al. A periplasmic polymer curves Vibrio cholerae and promotes pathogenesis. Cell. 2017;168(1–2):172-185.e15.
Zamorano-Sanchez D, Xian W, Lee CK, Salinas M, Thongsomboon W, Cegelski L, et al. Functional specialization in Vibrio cholerae diguanylate cyclases: distinct modes of motility suppression and c-di-GMP production. MBio. 2019;10(2):e00670-e719.
Yang S, Xi D, Wang X, Li Y, Li Y, Yan J, et al. Vibrio cholerae VC1741 (PsrA) enhances the colonization of the pathogen in infant mice intestines in the presence of the long-chain fatty acid, oleic acid. Microb Pathog. 2020;147:104443.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-DDCt method. Methods. 2001;25(4):402–8.
Xi D, Yang S, Liu Q, Li Y, Li Y, Yan J, et al. The response regulator ArcA enhances biofilm formation in the vpsT manner under the anaerobic condition in Vibrio cholerae. Microb Pathog. 2020;144:104197.
Liang H, Li L, Dong Z, Surette MG, Duan K. The YebC family protein PA0964 negatively regulates the pseudomonas aeruginosa quinolone signal system and pyocyanin production. J Bacteriol. 2008;190(18):6217–27.
Acknowledgements
This work was supported by the National Key Programs for Infectious Diseases of China (grant numbers 2017ZX10303405-001, 2017ZX10104002-001-006, 2018ZX1 0712001-017).
Funding
This work was supported by the National Key Programs for Infectious Diseases of China (grant numbers 2017ZX10303405-001, 2017ZX10104002-001–006, 2018ZX1 0712001–017). The funding bodies had no role in the design of the study, collection, analysis, and interpretation of data and in writing the manuscript.
Author information
Authors and Affiliations
Contributions
Yuehua Li: Investigation, Conceptualization, Project administration, Methodology, Writing—original draft. Junxiang Yan: Project administration, Methodology, Writing -original draft. Xueqian Guo: Data curation, Formal analysis. Xiaochen Wang: Methodology, Formal analysis. Fenxia Liu: Software, Visualization. Boyang Cao: Investigation, Conceptualization, Writing—original draft, Funding acquisition, Supervision, Writing—review & editing. The author(s) read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Li, Y., Yan, J., Guo, X. et al. The global regulators ArcA and CytR collaboratively modulate Vibrio cholerae motility. BMC Microbiol 22, 22 (2022). https://doi.org/10.1186/s12866-022-02435-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12866-022-02435-y