Abstract
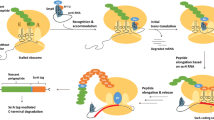
In Pseudomonas fluorescens CHA0, the synthesis of antifungal compounds is post-transcriptionally activated by the Gac/Rsm cascade. The two-component system GacS/GacA promotes transcription of three small regulatory RNAs (i.e., sRNAs), RsmX, RsmY, and RsmZ, which remove the regulatory proteins RsmA and RsmE from the ribosome-binding sites of exoproduct-related mRNAs. The GacS/GacA-dependent accumulation of RsmX/Y/Z and formation of RsmX/Y/Z-RsmA/E complexes relieve mRNA translational repression. Other bacteria as E. coli and Vibrio spp. utilize similar sRNA–protein based systems to adjust mRNA translation (e.g., the E. coli Csr system for carbon storage, motility and biofilm regulation). The Rsm/Csr sRNAs are remarkably similar in that they contain several stem-loops with an invariant GGA trinucleotide exposed in the hairpin loop that would be the characteristic structural-sequence motifs relevant for sRNA activity and stability. Here it is shown that the dysfunctional Gac/Rsm cascade of P. fluorescens ΔrsmXYZ mutants could be restored by appropriate transcription levels of artificial genes encoding RNAs with unrelated primary sequence but with two or more hairpins displaying the RsmA/E binding motifs. The results support the hypothesis that the molecular mimicry of Rsm/Csr sRNAs is based on proper secondary structures that expose critical binding motifs irrespective of their overall sequence.




Similar content being viewed by others
Abbreviations
- Gac:
-
Global antibiotic and cyanide control
- Rsm:
-
Regulator of secondary metabolism
- sRNAs:
-
Small non-coding regulatory RNAs
- arsm :
-
Artificial rsm-like RNA gene
References
Abramoff MD, Magelhaes PJ, Ram SJ (2004) Image processing with ImageJ. Biophotonics Int 11:36–42
Altier C, Suyemoto M, Ruiz AI, Burnham KD, Maurer R (2000) Characterization of two novel regulatory genes affecting Salmonella invasion gene expression. Mol Microbiol 35:635–646
Álvarez Crespo MC, Valverde C (2009) A single mutation in the oprF mRNA leader confers strict translational control by the Gac/Rsm system in Pseudomonas fluorescens CHA0. Curr Microbiol 58:182–188
Babitzke P, Romeo T (2007) CsrB sRNA family: sequestration of RNA-binding regulatory proteins. Curr Opin Microbiol 10:156–163
Baker CS, Morozov I, Suzuki K, Romeo T, Babitzke P (2002) CsrA regulates glycogen biosynthesis by preventing translation of glgC in Escherichia coli. Mol Microbiol 44:1599–1610
Baker CS, Eory LA, Yakhnin H, Mercante J, Romeo T, Babitzke P (2007) CsrA inhibits translation initiation of Escherichia coli hfq by binding to a single site overlapping the Shine-Dalgarno sequence. J Bacteriol 189:5472–5481
Bao Y, Lies DP, Fu H, Roberts GP (1991) An improved Tn7-based system for the single-copy insertion of cloned genes into chromosomes of gram-negative bacteria. Gene 109:167–168
Blumer C, Heeb S, Pessi G, Haas D (1999) Global GacA-steered control of cyanide and exoprotease production in Pseudomonas fluorescens involves specific ribosome binding sites. Proc Natl Acad Sci USA 96:14073–14078
Brennan RG, Link TM (2007) Hfq structure, function and ligand binding. Curr Opin Microbiol 10:125–133
Del Sal G, Manfioletti G, Schneider C (1988) A one-tube plasmid DNA mini-preparation suitable for sequencing. Nucleic Acids Res 16:9878
Dubey AK et al (2003) CsrA regulates translation of the Escherichia coli carbon starvation gene, cstA, by blocking ribosome access to the cstA transcript. J Bacteriol 185:4450–4460
Dubey AK, Baker CS, Romeo T, Babitzke P (2005) RNA sequence and secondary structure participate in high-affinity CsrA-RNA interaction. Rna 11:1579–1587
Egan SV, Yeoh HH, Bradbury JH (1998) Simple picrate paper kit for determination of the cyanogenic potential of cassava flour. J Sci Food Agric 76:39–48
Gamper M, Ganter B, Polito MR, Haas D (1992) RNA processing modulates the expression of the arcDABC operon in Pseudomonas aeruginosa. J Mol Biol 226:943–957
Heeb S, Blumer C, Haas D (2002) Regulatory RNA as mediator in GacA/RsmA-dependent global control of exoproduct formation in Pseudomonas fluorescens CHA0. J Bacteriol 184:1046–1056
Heeb S et al (2006) Functional analysis of the post-transcriptional regulator RsmA reveals a novel RNA-binding site. J Mol Biol 355:1026–1036
Jonas K, Edwards AN, Simm R, Romeo T, Romling U, Melefors O (2008) The RNA binding protein CsrA controls cyclic di-GMP metabolism by directly regulating the expression of GGDEF proteins. Mol Microbiol 70:236–257
Kay E, Dubuis C, Haas D (2005) Three small RNAs jointly ensure secondary metabolism and biocontrol in Pseudomonas fluorescens CHA0. Proc Natl Acad Sci USA 102:17136–17141
Kulkarni PR, Cui X, Williams JW, Stevens AM, Kulkarni RV (2006) Prediction of CsrA-regulating small RNAs in bacteria and their experimental verification in Vibrio fischeri. Nucleic Acids Res 34:3361–3369
Lapouge K et al (2007) Mechanism of hcnA mRNA recognition in the Gac/Rsm signal transduction pathway of Pseudomonas fluorescens. Mol Microbiol 66:341–356
Lapouge K, Schubert M, Allain FH, Haas D (2008) Gac/Rsm signal transduction pathway of gamma-proteobacteria: from RNA recognition to regulation of social behaviour. Mol Microbiol 67:241–253
Lenz DH, Miller MB, Zhu J, Kulkarni RV, Bassler BL (2005) CsrA and three redundant small RNAs regulate quorum sensing in Vibrio cholerae. Mol Microbiol 58:1186–1202
Liu MY et al (1997) The RNA molecule CsrB binds to the global regulatory protein CsrA and antagonizes its activity in Escherichia coli. J Biol Chem 272:17502–17510
Ma W, Cui Y, Liu Y, Dumenyo CK, Mukherjee A, Chatterjee AK (2001) Molecular characterization of global regulatory RNA species that control pathogenicity factors in Erwinia amylovora and Erwinia herbicola pv. gypsophilae. J Bacteriol 183:1870–1880
Majdalani N, Vanderpool CK, Gottesman S (2005) Bacterial small RNA regulators. Crit Rev Biochem Mol Biol 40:93–113
Maurhofer M, Reimmann C, Schmidli-Sacherer P, Heeb S, Haas D, Défago G (1998) Salicylic acid biosynthetic genes expressed in Pseudomonas fluorescens strain P3 improve the induction of systemic resistence in tobacco against tobacco necrosis virus. Phytopathology 88:678–684
Miller JH (1972) Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Ongena M et al (1999) Protection of cucumber against Pythium root rot by fluorescent pseudomonads: predominant role of induced resistance over siderophores and antibiosis. Plant Pathology 48:66–76
Reimmann C, Valverde C, Kay E, Haas D (2005) Posttranscriptional repression of GacS/GacA-controlled genes by the RNA-binding protein RsmE acting together with RsmA in the biocontrol strain Pseudomonas fluorescens CHA0. J Bacteriol 187:276–285
Romby P, Vandenesch F, Wagner EG (2006) The role of RNAs in the regulation of virulence-gene expression. Curr Opin Microbiol 9:229–236
Romeo T (1998) Global regulation by the small RNA-binding protein CsrA and the non-coding RNA molecule CsrB. Mol Microbiol 29:1321–1330
Romeo T, Gong M (1993) Genetic and physical mapping of the regulatory gene csrA on the Escherichia coli K-12 chromosome. J Bacteriol 175:5740–5741
Sacherer P, Defago G, Haas D (1994) Extracellular protease and phospholipase C are controlled by the global regulatory gene gacA in the biocontrol strain Pseudomonas fluorescens CHA0. FEMS Microbiol Lett 116:155–160
Sambrook J, Fritsch E, Maniatis T (1989) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, New York
Schubert M et al (2007) Molecular basis of messenger RNA recognition by the specific bacterial repressing clamp RsmA/CsrA. Nat Struct Mol Biol 14:807–813
Siddiqui IA, Haas D, Heeb S (2005) Extracellular protease of Pseudomonas fluorescens CHA0, a biocontrol factor with activity against the root-knot nematode Meloidogyne incognita. Appl Environ Microbiol 71:5646–5649
Sorger-Domenigg T, Sonnleitner E, Kaberdin VR, Blasi U (2007) Distinct and overlapping binding sites of Pseudomonas aeruginosa Hfq and RsmA proteins on the non-coding RNA RsmY. Biochem Biophys Res Commun 352:769–773
Storz G, Altuvia S, Wassarman KM (2005) An abundance of RNA regulators. Annu Rev Biochem 74:199–217
Suzuki K, Babitzke P, Kushner SR, Romeo T (2006) Identification of a novel regulatory protein (CsrD) that targets the global regulatory RNAs CsrB and CsrC for degradation by RNase E. Genes Dev 20:2605–2617
Valverde C, Haas D (2008) Small RNAs controlled by two component systems. Chapter 5. In: Utsumi R (ed) Bacterial signal transduction: network and drug targets, 2nd edn. Landes Bioscience, Austin
Valverde C, Heeb S, Keel C, Haas D (2003) RsmY, a small regulatory RNA, is required in concert with RsmZ for GacA-dependent expression of biocontrol traits in Pseudomonas fluorescens CHA0. Mol Microbiol 50:1361–1379
Valverde C, Lindell M, Wagner EG, Haas D (2004) A repeated GGA motif is critical for the activity and stability of the riboregulator RsmY of Pseudomonas fluorescens. J Biol Chem 279:25066–25074
Voisard C, Bull CT, Keel C, Laville J, Maurhofer M, Schnider M (1994) Biocontrol of root diseases by Pseudomonas fluorescens CHA0: current concepts and experimental approaches. In: O’Gara F, Dowling DN, Boesten B (eds) Molecular ecology of rhizosphere microorganisms. VCH Verlagsgesellschaft mbH, pp 67–89
Wang X, Dubey AK, Suzuki K, Baker CS, Babitzke P, Romeo T (2005) CsrA post-transcriptionally represses pgaABCD, responsible for synthesis of a biofilm polysaccharide adhesin of Escherichia coli. Mol Microbiol 56:1648–1663
Wei BL, Brun-Zinkernagel AM, Simecka JW, Pruss BM, Babitzke P, Romeo T (2001) Positive regulation of motility and flhDC expression by the RNA-binding protein CsrA of Escherichia coli. Mol Microbiol 40:245–256
Weilbacher T et al (2003) A novel sRNA component of the carbon storage regulatory system of Escherichia coli. Mol Microbiol 48:657–670
Zuber S et al (2003) GacS sensor domains pertinent to the regulation of exoproduct formation and to the biocontrol potential of Pseudomonas fluorescens CHA0. Mol Plant Microbe Interact 16:634–644
Zuker M (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31:3406–3415
Acknowledgments
This work was supported by grants from Banco RIO, CONICET and Universidad Nacional de Quilmes (Argentina). The author is grateful to Prof. Dieter Haas (DMF, Université de Lausanne, Switzerland) for providing P. fluorescens strains. The author would like to acknowledge the critical comments of two anonymous reviewers which contributed to improve this manuscript. C. V. is member of CONICET (Argentina).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by John Helmann.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Figure S1:
Stability of RsmY-like RNA homologues expressed in P. fluorescens. (JPG 183 kb)
Supplementary Figure S2:
Biological activity of RsmY-like RNA homologues. (JPG 293 kb)
Supplementary file 1:
Sequence of studied RNA molecules and comparative analysis of CsrA/RsmA recognition motifs. (DOC 76 kb)
Supplementary file 2:
Nucleotide substitutions in hairpins of RsmZ RNA tend to preserve secondary structure. (DOC 220 kb)
Rights and permissions
About this article
Cite this article
Valverde, C. Artificial sRNAs activating the Gac/Rsm signal transduction pathway in Pseudomonas fluorescens . Arch Microbiol 191, 349–359 (2009). https://doi.org/10.1007/s00203-009-0459-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-009-0459-x