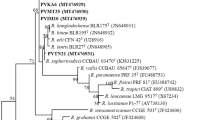
Abstract
Great genetic diversity was revealed among 75 rhizobal isolates associated with Vicia faba grown in Chinese fields with AFLP, ARDRA, 16S rDNA sequencing, DNA–DNA hybridization, BOX-PCR and RFLP of PCR-amplified nodD and nodC. Most of the isolates were Rhizobium leguminosarum, and six isolates belonged to an unnamed Rhizobium species. In the homogeneity analysis, the isolates were grouped into three clusters corresponding to (1) autumn sowing (subtropical) region where the winter ecotype of V. faba was cultivated, (2) spring sowing (temperate) region where the spring ecotype was grown, and (3) Yunnan province where the intermediate ecotype was sown either in spring or in autumn. Nonrandom associations were found among the nod genotypes, genomic types and ecological regions, indicating an epidemic symbiotic gene transfer pattern among different genomic backgrounds within an ecological region and a relatively limited transfer pattern between different regions. Conclusively, the present results suggested an endemic population structure of V. faba rhizobia in Chinese fields and demonstrated a novel rhizobium associated with faba bean.





Similar content being viewed by others
References
Andronov EE, Terefework Z, Roumiantseva ML, Dzyubenko NI, Onichtchouk OP, Kurchak ON, Dresler-Nurmi A, Young JPW, Simarov BV, Lindrström K (2003) Symbiotic and genetic diversity of Rhizobium galegae isolates collected from the Galega orientalis gene center in the Caucasus. Appl Environ Microbiol 69:1067–1074
Bond DA, Lawes DA, Hawtin GC, Saxena MC, Stephens JH (1985) Faba bean (Vicia faba L.). In: Summerfield RJ, Roberts EH (eds) Grain legume crops Collins professional and technical books. William Collins Sons & Co. Ltd, London, pp 199–265
Cho JC, Tiedje JM (2000) Biogeography and degree of endemicity of fluorescent Pseudomonas strains in soil. Appl Environ Microbiol 66:5448–5456
De Ley J (1970) Reexamination of the association between melting point, buoyant density, and chemical base composition of DNA. J Bacteriol 101:738–754
De Ley J, Cattoir H, Reynaerts A (1970) The quantitative measurement of DNA hybridization from renaturation rates. Eur J Biochem 12:133–142
Duke JA (1981) Handbook of legumes of world economic importance. Plenum Press, New York, pp 275–279
George RAT (1999) Vegetable seed production, 2nd edn. CABI Publishing, Wallingford, pp 204–205
Graham PH, Vance CP (2000) Nitrogen fixation in perspective: an overview of research and extension needs. Field Crops Res 65:93–106
Graham PH, Vance CP (2003) Legumes: importance and constraints to greater use. Plant Physiol 131:872–877
Healy M, Huong J, Bittner T, Lising M, Frye S, Raza S, Schrock R, Manry J, Renwick A, Nieto R, Woods C, Versalovic J, Lupski JR (2005) Microbial DNA typing by automated repetitive-sequence-based PCR. J Clin Microbiol 43:199–207
Howieson JG, O’Hara GW, Carr SJ (2000) Changing roles for legumes in Mediterranean agriculture: developments from an Australian perspective. Field Crops Res 65:107–122
Jeanmougin F, Thompson JD, Gouy M, Higgins DG, Gibson TJ (1998) Multiple sequence alignment with Clustal x. Trends Biochem Sci 23:403–405
Jordan DC (1984) Genus I. Rhizobium Frank 1889, 338AL. In: Krieg NR, Holt G (eds) Bergey’s manual of systematic bacteriology, vol 1. Williams and Wilkins, Baltimore, pp 235–242
Koeuth T, Versalovic J, Lupski JR (1995) Differential subsequence conservation of interspersed repetitive Streptococcus pneumoniae BOX elements in diverse bacteria. Genome Res 5:408–418
Kumar S, Tamura K, Nei M (2004) MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5:150–163
Laguerre G, Mavingui P, Allard MR, Charnay MP, Louvrier P, Mazurier SI, Rigottier-Gois L, Amarger N (1996) Typing of rhizobia by PCR DNA fingerprinting and PCR-restriction fragment length polymorphism analysis of chromosomal and symbiotic gene regions: application to Rhizobium leguminosarum and its different biovars. Appl Environ Microbiol 62:2029–2036
Laguerre G, Louvrier P, Allard MR, Amarger N (2003) Compatibility of rhizobial genotypes within natural populations of Rhizobium leguminosarum biovar viciae for nodulation of host legumes. Appl Environ Microbiol 69:2276–2283
Liu J, Wang ET, Chen WX (2005) Diverse rhizobia associated with woody legumes Wisteria sinensis, Cercis racemosa and Amorpha fruticosa grown in the temperate zone of China. Syst Appl Microbiol 28:465–477
Lohrke SM, Orf JH, Sadowsky MJ (1996) Inheritance of host controlled restriction of nodulation by Bradyrhizobium japonicum strain USDA 110. Crop Sci 36:1271–1277
Louvrier P, Laguerre G, Amarger N (1996) Distribution of symbiotic genotypes in Rhizobium leguminosarum biovar viciae populations isolated directly from soils. Appl Environ Microbiol 62:4202–4205
Marmur J (1961) A procedure for the isolation of DNA from microorganisms. J Mol Biol 3:208–218
Mutch LA, Tamimi SM, Young JPW (2003) Genotypic characterisation of rhizobia nodulating Vicia faba from the soils of Jordan: a comparison with UK isolates. Soil Biol Biochem 35:709–714
Mutch LA, Young JPW (2004) Diversity and specificity of Rhizobium leguminosarum biovar viciae on wild and cultivated legumes. Mol Ecol 13:2435–2444
Nick G, Rasanen LA, de Lajudie P, Gillis M, Lindström K (1999) Genomic screening of rhizobia isolated from root nodules of tropical leguminous trees using DNA-DNA dot-blot hybridization and rep-PCR. Syst Appl Microbiol 22:287–299
Rademaker JLW, Hoste B, Louws FJ, Kersters K, Swings J, Vauterin L, Vauterin P, de Bruijn FJ (2000) Comparison of AFLP and rep-PCR genomic fingerpring with DNA-DNA homology studies: Xanthomonas as a model system. Int J Syst Evol Microbiol 50:665–677
Resh SC, Binkley D, Parrotta JA (2002) Greater soil carbon sequestration under nitrogen-fixing trees compared with Eucalyptus species. Ecosystem 5:217–231
Sarita S, Sharma PK, Priefer UB, Prell J (2005) Direct amplification of rhizobial nodC sequences from soil total DNA and comparison to nodC diversity of root nodule isolates. FEMS Microbiol Ecol 54:1–11
Souza V, Eguiarte LE (1997) Bacteria gone native vs. bacteria gone awry? Plasmidic transfer and bacterial evolution. Proc Natl Acad Sci USA 94:5501–5503
Terefework Z, Kaijalainen S, Lindström K (2001) AFLP fingerprinting as a tool to study the genetic diversity of Rhizobium galegae isolated from Galega orientalis and Galega officinalis. J Biotechnol 91:169–180
Thompson JD, Higgins DG, Gibson TJ (1994) Clustal-W—improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Thompson HC, Kelly WC (1957) Vegetable crops, 5th edn. Mograw-Hill Book Company, New York
van Berkum P, Beyene D, Vera FT, Keyser HH (1995) Variability among Rhizobium strains originating from nodules of Vicia faba. Appl Environ Microbiol 61:2649–2653
Vincent JM (1970) A manual for the practical study of root nodule bacteria. IBP handbook 15. Blackwell, Oxford
Vinuesa P, Rademaker JL, Bruijn FJ, Werner D (1998) Genotypic characterization of Bradyrhizobium strains nodulating endemic woody legumes of the Canary islands by PCR-restriction fragment length polymorphism analysis of genes encoding 16S rRNA (16S rDNA) and 16S-23S rDNA intergenic spacers, repetitive extragenic palindromic PCR genomic fingerprinting, and partial 16S rDNA sequencing. Appl Environ Microbiol 64:2096–2104
Weir BS, Turner SJ, Silvester WB, Park DC, Young JM (2004) Unexpectedly diverse Mesorhizobium strains and Rhizobium leguminosarum nodulate native legume genera of New Zealand, while introduced legume weeds are nodulated by Bradyrhizobium species. Appl Environ Microbiol 70:5980–5987
Weisburg WG, Barns SM, Pelletior DA, Lane DJ (1991) 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173:697–703
Wolde-meskel E, Terefework Z, Lindström K, Frostegård Å (2004) Rhizobia nodulating African Acacia spp. and Sesbania sesban trees in southern Ethiopian soils are metabolically and genomically diverse Soil Biol Biochem 36:2013–2025
Ye Y (2003) “Zhong Guo Can Dou Xue” (Chinese faba bean) (in Chinese). China Agriculture Press, Beijing
Young JPW, Mutch LA, Ashford DA, Zézé A, Mutch KE (2003) The molecular evolution of host specificity in the rhizobium-legume symbiosis. In: Hails R, Godfray HCJ, Beringer J (eds) Genes in the environment. Blackwell, Oxford, pp 245–257
Zézé A, Mutch LA, Young JPW (2001) Direct amplification of nodD from community DNA reveals the genetic diversity of Rhizobium leguminosarum in soil. Environ Microbiol 3:363–370
Acknowledgments
We thank Dr. Min Qi Lu, Dr. Qiang Chen and other collaborators for their valuable contribution in collecting and isolating the rhizobia. This work was financial supported by the foundation of the State Key Basic Research and Development Plan of China (grant 2006CB100206), and by National Project for Basic S&T Platform Construction (grant 2005DKA21201-10).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Erko Stackebrandt.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Tian, C.F., Wang, E.T., Han, T.X. et al. Genetic diversity of rhizobia associated with Vicia faba in three ecological regions of China. Arch Microbiol 188, 273–282 (2007). https://doi.org/10.1007/s00203-007-0245-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-007-0245-6