Abstract
Summary
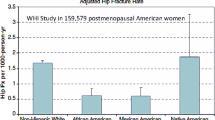
Whether the PGS developed using data from European ancestry is predictive of fracture risk for minorities remains unclear. This study demonstrated that PGSs based on common BMD-related genetic variants discovered in the European ancestry cohort are predictive of fracture risk in people of Asian but not African ancestry.
Purpose
Large-scale genome-wide association studies (GWAS) on bone mineral density (BMD) have been conducted predominantly in European cohorts. Genetic models based on common variants associated with BMD have been evaluated using almost exclusively European data, which could potentially exacerbate health disparities due to different linkage disequilibrium among different ethnic groups.
Methods
UK Biobank (UKB) is a large-scale population-based observational study starting in 2006 that recruited 502,617 individuals aged between 40 and 69 years with genotypic and phenotypic data available. Based on the summary statistics of two GWAS studies of femoral neck BMD and total body BMD, we derived four PGSs and assessed the association between each PGS and prevalent/incident fractures within each ethnic group separately using Multivariate logistic regressions and Cox proportional hazard models. All models were adjusted for age, sex, and the first four principal components.
Results
We assessed four PGSs derived from European cohorts. Significant associations were observed between PGSs and fracture in European and Asian cohorts but not in the African cohort. Of all four PGSs, \({\mathrm{PGS}\_\mathrm{TB}}_{\mathrm{ldpred}}\) performed the best. A standard deviation decreases in \({\mathrm{PGS}\_\mathrm{TB}}_{\mathrm{ldpred}}\) were associated with an increased hazard ratio (HR) of 1.24 (1.22–1.27), 1.28 (0.83–1.99), and 1.34 (1.10–1.64) in European, African, and Asian ancestry, respectively. A low BMD-related PGS is associated with up to 2.35- and 4.31-fold increased fracture risk in European and Asian populations.
Conclusions
These results showed that PGSs based on common BMD-related genetic variants discovered in the European ancestry cohort are predictive of fracture risk in people of Asian but not African ancestry.



Similar content being viewed by others
Data availability
Data sharing does not apply to this article as no datasets were generated during the current study.
Abbreviations
- BMD:
-
Bone mineral density
- PGS:
-
Polygenic score
- GWAS:
-
Genome-wide association study
- FRAX:
-
Fracture risk assessment tool
- SNP:
-
Single nucleotide polymorphisms
References
Johnell O, Kanis JA (2006) An estimate of the worldwide prevalence and disability associated with osteoporotic fractures. Osteoporos Int 17:1726–1733. https://doi.org/10.1007/s00198-006-0172-4
Slemenda CW, Turner CH, Peacock M, Christian JC, Sorbel J, Hui SL, Johnston CC (1996) The genetics of proximal femur geometry, distribution of bone mass and bone mineral density. Osteoporosis Int 6:178–182
Smith DM, Nance WE, Kang KW, Christian JC, Johnston CC (1973) Genetic factors in determining bone mass. J Clin Invest 52:2800–2808
Guéguen R, Jouanny P, Guillemin F, Kuntz C, Pourel J, Siest G (1995) Segregation analysis and variance components analysis of bone mineral density in healthy families. J Bone Miner Res 10:2017–2022
Krall EA, Dawson-Hughes B (1993) Heritable and life-style determinants of bone mineral density. J Bone Miner Res 8:1–9
Ralston SH, De Crombrugghe B (2006) Genetic regulation of bone mass and susceptibility to osteoporosis. Genes Dev 20:2492–2506
Zhu X, Bai W, Zheng H (2021) Twelve years of GWAS discoveries for osteoporosis and related traits: advances, challenges and applications. Bone Res 9:23. https://doi.org/10.1038/s41413-021-00143-3
Medina-Gomez C, Kemp JP, Trajanoska K, Luan J, Chesi A, Ahluwalia TS, Mook-Kanamori DO, Ham A, Hartwig FP, Evans DS, Joro R, Nedeljkovic I, Zheng HF, Zhu K, Atalay M, Liu CT, Nethander M, Broer L, Porleifsson G, Mullin BH, Handelman SK, Nalls MA, Jessen LE, Heppe DHM, Richards JB, Wang C, Chawes B, Schraut KE, Amin N, Wareham N, Karasik D, Van der Velde N, Ikram MA, Zemel BS, Zhou Y, Carlsson CJ, Liu Y, McGuigan FE, Boer CG, Bønnelykke K, Ralston SH, Robbins JA, Walsh JP, Zillikens MC, Langenberg C, Li-Gao R, Williams FMK, Harris TB, Akesson K, Jackson RD, Sigurdsson G, den Heijer M, van der Eerden BCJ, van de Peppel J, Spector TD, Pennell C, Horta BL, Felix JF, Zhao JH, Wilson SG, de Mutsert R, Bisgaard H, Styrkársdóttir U, Jaddoe VW, Orwoll E, Lakka TA, Scott R, Grant SFA, Lorentzon M, van Duijn CM, Wilson JF, Stefansson K, Psaty BM, Kiel DP, Ohlsson C, Ntzani E, van Wijnen AJ, Forgetta V, Ghanbari M, Logan JG, Williams GR, Bassett JHD, Croucher PI, Evangelou E, Uitterlinden AG, Ackert-Bicknell CL, Tobias JH, Evans DM, Rivadeneira F (2018) Life-course genome-wide association study meta-analysis of total body BMD and assessment of age-specific effects. Am J Hum Genet 102:88–102
Estrada K, Styrkarsdottir U, Evangelou E, Hsu Y, Duncan EL, Ntzani EE, Oei L, Albagha OME, Amin N, Kemp JP, Koller DL, Li G, Liu C, Minster RL, Moayyeri A, Vandenput L, Willner D, Xiao S, Yerges-Armstrong L, Zheng H, Alonso N, Eriksson J, Kammerer CM, Kaptoge SK, Leo PJ, Thorleifsson G, Wilson SG, Wilson JF, Aalto V, Alen M, Aragaki AK, Aspelund T, Center JR, Dailiana Z, Duggan DJ, Garcia M, Garcia-Giralt N, Giroux S, Hallmans G, Hocking LJ, Husted LB, Jameson KA, Khusainova R, Kim GS, Kooperberg C, Koromila T, Kruk M, Laaksonen M, Lacroix AZ, Lee SH, Leung PC, Lewis JR, Masi L, Mencej-Bedrac S, Nguyen TV, Nogues X, Patel MS, Prezelj J, Rose LM, Scollen S, Siggeirsdottir K, Smith AV, Svensson O, Trompet S, Trummer O, van Schoor NM, Woo J, Zhu K, Balcells S, Brandi ML, Buckley BM, Cheng S, Christiansen C, Cooper C, Dedoussis G, Ford I, Frost M, Goltzman D, González-Macías J, Kähönen M, Karlsson M, Khusnutdinova E, Koh J, Kollia P, Langdahl BL, Leslie WD, Lips P, Ljunggren Ö, Lorenc RS, Marc J, Mellström D, Obermayer-Pietsch B, Olmos JM, Pettersson-Kymmer U, Reid DM, Riancho JA, Ridker PM, Rousseau F, Slagboom PE, Tang NLS, Urreizti R, Van Hul W, Viikari J, Zarrabeitia MT, Aulchenko YS, Castano-Betancourt M, Grundberg E, Herrera L, Ingvarsson T, Johannsdottir H, Kwan T, Li R, Luben R, Medina-Gómez C, Palsson ST, Reppe S, Rotter JI, Sigurdsson G, van Meurs JBJ, Verlaan D, Williams FMK, Wood AR, Zhou Y, Gautvik KM, Pastinen T, Raychaudhuri S, Cauley JA, Chasman DI, Clark GR, Cummings SR, Danoy P, Dennison EM, Eastell R, Eisman JA, Gudnason V, Hofman A, Jackson RD, Jones G, Jukema JW, Khaw K, Lehtimäki T, Liu Y, Lorentzon M, McCloskey E, Mitchell BD, Nandakumar K, Nicholson GC, Oostra BA, Peacock M, Pols HAP, Prince RL, Raitakari O, Reid IR, Robbins J, Sambrook PN, Sham PC, Shuldiner AR, Tylavsky FA, van Duijn CM, Wareham NJ, Cupples LA, Econs MJ, Evans DM, Harris TB, Kung AWC, Psaty BM, Reeve J, Spector TD, Streeten EA, Zillikens MC, Thorsteinsdottir U, Ohlsson C, Karasik D, Richards JB, Brown MA, Stefansson K, Uitterlinden AG, Ralston SH, Ioannidis JPA, Kiel DP, Rivadeneira F (2012) Genome-wide meta-analysis identifies 56 bone mineral density loci and reveals 14 loci associated with risk of fracture. Nat Genet 44:491–501. https://doi.org/10.1038/ng.2249
Lewis CM, Vassos E (2020) Polygenic risk scores: from research tools to clinical instruments. Genome Med 12:44. https://doi.org/10.1186/s13073-020-00742-5
Abraham G, Havulinna AS, Bhalala OG, Byars SG, De Livera AM, Yetukuri L, Tikkanen E, Perola M, Schunkert H, Sijbrands EJ, Palotie A, Samani NJ, Salomaa V, Ripatti S, Inouye M (2016) Genomic prediction of coronary heart disease. Eur Heart J 37:3267–3278
Mavaddat N, Pharoah PD, Michailidou K, Tyrer J, Brook MN, Bolla MK, Wang Q, Dennis J, Dunning AM, Shah M, Luben R, Brown J, Bojesen SE, Nordestgaard BG, Nielsen SF, Flyger H, Czene K, Darabi H, Eriksson M, Peto J, Dos-Santos-Silva I, Dudbridge F, Johnson N, Schmidt MK, Broeks A, Verhoef S, Rutgers EJ, Swerdlow A, Ashworth A, Orr N, Schoemaker MJ, Figueroa J, Chanock SJ, Brinton L, Lissowska J, Couch FJ, Olson JE, Vachon C, Pankratz VS, Lambrechts D, Wildiers H, Van Ongeval C, van Limbergen E, Kristensen V, Grenaker Alnæs G, Nord S, Borresen-Dale AL, Nevanlinna H, Muranen TA, Aittomäki K, Blomqvist C, Chang-Claude J, Rudolph A, Seibold P, Flesch-Janys D, Fasching PA, Haeberle L, Ekici AB, Beckmann MW, Burwinkel B, Marme F, Schneeweiss A, Sohn C, Trentham-Dietz A, Newcomb P, Titus L, Egan KM, Hunter DJ, Lindstrom S, Tamimi RM, Kraft P, Rahman N, Turnbull C, Renwick A, Seal S, Li J, Liu J, Humphreys K, Benitez J, Pilar Zamora M, Arias Perez JI, Menéndez P, Jakubowska A, Lubinski J, Jaworska-Bieniek K, Durda K, Bogdanova NV, Antonenkova NN, Dörk T, Anton-Culver H, Neuhausen SL, Ziogas A, Bernstein L, Devilee P, Tollenaar RA, Seynaeve C, van Asperen CJ, Cox A, Cross SS, Reed MW, Khusnutdinova E, Bermisheva M, Prokofyeva D, Takhirova Z, Meindl A, Schmutzler RK, Sutter C, Yang R, Schürmann P, Bremer M, Christiansen H, Park-Simon TW, Hillemanns P, Guénel P, Truong T, Menegaux F, Sanchez M, Radice P, Peterlongo P, Manoukian S, Pensotti V, Hopper JL, Tsimiklis H, Apicella C, Southey MC, Brauch H, Brüning T, Ko YD, Sigurdson AJ, Doody MM, Hamann U, Torres D, Ulmer HU, Försti A, Sawyer EJ, Tomlinson I, Kerin MJ, Miller N, Andrulis IL, Knight JA, Glendon G, Marie Mulligan A, Chenevix-Trench G, Balleine R, Giles GG, Milne RL, McLean C, Lindblom A, Margolin S, Haiman CA, Henderson BE, Schumacher F, Le Marchand L, Eilber U, Wang-Gohrke S, Hooning MJ, Hollestelle A, van den Ouweland AM, Koppert LB, Carpenter J, Clarke C, Scott R, Mannermaa A, Kataja V, Kosma VM, Hartikainen JM, Brenner H, Arndt V, Stegmaier C, Karina Dieffenbach A, Winqvist R, Pylkäs K, Jukkola-Vuorinen A, Grip M, Offit K, Vijai J, Robson M, Rau-Murthy R, Dwek M, Swann R, Annie Perkins K, Goldberg MS, Labrèche F, Dumont M, Eccles DM, Tapper WJ, Rafiq S, John EM, Whittemore AS, Slager S, Yannoukakos D, Toland AE, Yao S, Zheng W, Halverson SL, González-Neira A, Pita G, Rosario Alonso M, Álvarez N, Herrero D, Tessier DC, Vincent D, Bacot F, Luccarini C, Baynes C, Ahmed S, Maranian M, Healey CS, Simard J, Hall P, Easton DF, Garcia-Closas M (2015) Prediction of breast cancer risk based on profiling with common genetic variants. J Natl Cancer Inst 107:djv036. https://doi.org/10.1093/jnci/djv036
Mavaddat N, Michailidou K, Dennis J, Lush M, Fachal L, Lee A, Tyrer JP, Chen TH, Wang Q, Bolla MK, Yang X, Adank MA, Ahearn T, Aittomäki K, Allen J, Andrulis IL, Anton-Culver H, Antonenkova NN, Arndt V, Aronson KJ, Auer PL, Auvinen P, Barrdahl M, Beane Freeman LE, Beckmann MW, Behrens S, Benitez J, Bermisheva M, Bernstein L, Blomqvist C, Bogdanova NV, Bojesen SE, Bonanni B, Børresen-Dale AL, Brauch H, Bremer M, Brenner H, Brentnall A, Brock IW, Brooks-Wilson A, Brucker SY, Brüning T, Burwinkel B, Campa D, Carter BD, Castelao JE, Chanock SJ, Chlebowski R, Christiansen H, Clarke CL, Collée JM, Cordina-Duverger E, Cornelissen S, Couch FJ, Cox A, Cross SS, Czene K, Daly MB, Devilee P, Dörk T, Dos-Santos-Silva I, Dumont M, Durcan L, Dwek M, Eccles DM, Ekici AB, Eliassen AH, Ellberg C, Engel C, Eriksson M, Evans DG, Fasching PA, Figueroa J, Fletcher O, Flyger H, Försti A, Fritschi L, Gabrielson M, Gago-Dominguez M, Gapstur SM, García-Sáenz JA, Gaudet MM, Georgoulias V, Giles GG, Gilyazova IR, Glendon G, Goldberg MS, Goldgar DE, González-Neira A, Grenaker Alnæs GI, Grip M, Gronwald J, Grundy A, Guénel P, Haeberle L, Hahnen E, Haiman CA, Håkansson N, Hamann U, Hankinson SE, Harkness EF, Hart SN, He W, Hein A, Heyworth J, Hillemanns P, Hollestelle A, Hooning MJ, Hoover RN, Hopper JL, Howell A, Huang G, Humphreys K, Hunter DJ, Jakimovska M, Jakubowska A, Janni W, John EM, Johnson N, Jones ME, Jukkola-Vuorinen A, Jung A, Kaaks R, Kaczmarek K, Kataja V, Keeman R, Kerin MJ, Khusnutdinova E, Kiiski JI, Knight JA, Ko YD, Kosma VM, Koutros S, Kristensen VN, Krüger U, Kühl T, Lambrechts D, Le Marchand L, Lee E, Lejbkowicz F, Lilyquist J, Lindblom A, Lindström S, Lissowska J, Lo WY, Loibl S, Long J, Lubiński J, Lux MP, MacInnis RJ, Maishman T, Makalic E, Maleva Kostovska I, Mannermaa A, Manoukian S, Margolin S, Martens JWM, Martinez ME, Mavroudis D, McLean C, Meindl A, Menon U, Middha P, Miller N, Moreno F, Mulligan AM, Mulot C, Muñoz-Garzon VM, Neuhausen SL, Nevanlinna H, Neven P, Newman WG, Nielsen SF, Nordestgaard BG, Norman A, Offit K, Olson JE, Olsson H, Orr N, Pankratz VS, Park-Simon TW, Perez JIA, Pérez-Barrios C, Peterlongo P, Peto J, Pinchev M, Plaseska-Karanfilska D, Polley EC, Prentice R, Presneau N, Prokofyeva D, Purrington K, Pylkäs K, Rack B, Radice P, Rau-Murthy R, Rennert G, Rennert HS, Rhenius V, Robson M, Romero A, Ruddy KJ, Ruebner M, Saloustros E, Sandler DP, Sawyer EJ, Schmidt DF, Schmutzler RK, Schneeweiss A, Schoemaker MJ, Schumacher F, Schürmann P, Schwentner L, Scott C, Scott RJ, Seynaeve C, Shah M, Sherman ME, Shrubsole MJ, Shu XO, Slager S, Smeets A, Sohn C, Soucy P, Southey MC, Spinelli JJ, Stegmaier C, Stone J, Swerdlow AJ, Tamimi RM, Tapper WJ, Taylor JA, Terry MB, Thöne K, Tollenaar RAEM, Tomlinson I, Truong T, Tzardi M, Ulmer HU, Untch M, Vachon CM, van Veen EM, Vijai J, Weinberg CR, Wendt C, Whittemore AS, Wildiers H, Willett W, Winqvist R, Wolk A, Yang XR, Yannoukakos D, Zhang Y, Zheng W, Ziogas A, ABCTB Investigators, kConFab/AOCS Investigators, NBCS Collaborators, Dunning AM, Thompson DJ, Chenevix-Trench G, Chang-Claude J, Schmidt MK, Hall P, Milne RL, Pharoah PDP, Antoniou AC, Chatterjee N, Kraft P, García-Closas M, Simard J, Easton DF (2019) Polygenic risk scores for prediction of breast cancer and breast cancer subtypes. Am J Hum Genet 104:21–34
Khera AV, Chaffin M, Aragam KG, Haas ME, Roselli C, Choi SH, Natarajan P, Lander ES, Lubitz SA, Ellinor PT, Kathiresan S (2018) Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat Genet 50:1219–1224. https://doi.org/10.1038/s41588-018-0183-z
Belsky DW, Moffitt TE, Sugden K, Williams B, Houts R, McCarthy J, Caspi A (2013) Development and evaluation of a genetic risk score for obesity. Biodemography Soc Biol 59:85–100. https://doi.org/10.1080/19485565.2013.774628
Vassos E, Di Forti M, Coleman J, Iyegbe C, Prata D, Euesden J, O’Reilly P, Curtis C, Kolliakou A, Patel H, Newhouse S, Traylor M, Ajnakina O, Mondelli V, Marques TR, Gardner-Sood P, Aitchison KJ, Powell J, Atakan Z, Greenwood KE, Smith S, Ismail K, Pariante C, Gaughran F, Dazzan P, Markus HS, David AS, Lewis CM, Murray RM, Breen G (2017) An examination of polygenic score risk prediction in individuals with first-episode psychosis. Biol Psychiatry 81:470–477
Wang L, Desai H, Verma SS, Le A, Hausler R, Verma A, Judy R, Doucette A, Gabriel PE, Center RG, Nathanson KL, Damrauer SM, Mowery DL, Ritchie MD, Kember RL, Maxwell KN (2022) Performance of polygenic risk scores for cancer prediction in a racially diverse academic biobank. Genet Med 24:601–609
Martin AR, Kanai M, Kamatani Y, Okada Y, Neale BM, Daly MJ (2019) Clinical use of current polygenic risk scores may exacerbate health disparities. Nat Genet 51:584–591. https://doi.org/10.1038/s41588-019-0379-x
Eriksson J, Evans DS, Nielson CM, Shen J, Srikanth P, Hochberg M, McWeeney S, Cawthon PM, Wilmot B, Zmuda J, Tranah G, Mirel DB, Challa S, Mooney M, Crenshaw A, Karlsson M, Mellström D, Vandenput L, Orwoll E, Ohlsson C (2015) Limited clinical utility of a genetic risk score for the prediction of fracture risk in elderly subjects. J Bone Miner Res 30:184–194. https://doi.org/10.1002/jbmr.2314
Warrington NM, Kemp JP, Tilling K, Tobias JH, Evans DM (2015) Genetic variants in adult bone mineral density and fracture risk genes are associated with the rate of bone mineral density acquisition in adolescence. Hum Mol Genet 24:4158–4166. https://doi.org/10.1093/hmg/ddv143
Nethander M, Pettersson-Kymmer U, Vandenput L, Lorentzon M, Karlsson M, Mellström D, Ohlsson C (2020) BMD-related genetic risk scores predict site-specific fractures as well as trabecular and cortical bone microstructure. J Clin Endocrinol Metab 105:e1344–e1357. https://doi.org/10.1210/clinem/dgaa082
Yau MS, Kuipers AL, Price R, Nicolas A, Tajuddin SM, Handelman SK, Arbeeva L, Chesi A, Hsu YH, Liu CT, Karasik D, Zemel BS, Grant SF, Jordan JM, Jackson RD, Evans MK, Harris TB, Zmuda JM, Kiel DP (2021) A meta-analysis of the transferability of bone mineral density genetic loci associations from European to African ancestry populations. J Bone Miner Res 36:469–479. https://doi.org/10.1002/jbmr.4220
Xiao X, Wu Q (2020) Association between a literature-based genetic risk score and bone mineral density of African American women in Women Health Initiative Study. Osteoporosis Int 31:913–920. https://doi.org/10.1007/s00198-019-05244-8
Styrkarsdottir U, Halldorsson BV, Gudbjartsson DF, Tang NLS, Koh J, Xiao S, Kwok TCY, Kim GS, Chan JCN, Cherny S, Lee SH, Kwok A, Ho S, Gretarsdottir S, Pop Kostic J, Palsson ST, Sigurdsson G, Sham PC, Kim B, Kung AWC, Kim S, Woo J, Leung P, Kong A, Thorsteinsdottir U, Stefansson K (2010) European bone mineral density loci are also associated with BMD in East-Asian populations. PLoS ONE 5:e13217
Cauley JA (2011) Defining ethnic and racial differences in osteoporosis and fragility fractures. Clin Orthop Relat Res 469(7):1891–9. https://doi.org/10.1007/s11999-011-1863-5
Lewiecki EM, Wright NC, Singer AJ (2020) Racial disparities, FRAX, and the care of patients with osteoporosis. Osteoporosis Int 31:2069–2071. https://doi.org/10.1007/s00198-020-05655-y
Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J, Downey P, Elliott P, Green J, Landray M, Liu B, Matthews P, Ong G, Pell J, Silman A, Young A, Sprosen T, Peakman T, Collins R (2015) UK Biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med 12:e1001779
Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, Motyer A, Vukcevic D, Delaneau O, O’Connell J, Cortes A, Welsh S, Young A, Effingham M, McVean G, Leslie S, Allen N, Donnelly P, Marchini J (2018) The UK Biobank resource with deep phenotyping and genomic data. Nature 562:203–209. https://doi.org/10.1038/s41586-018-0579-z
McCarthy S, Das S, Kretzschmar W, Delaneau O, Wood AR, Teumer A, Kang HM, Fuchsberger C, Danecek P, Sharp K, Luo Y, Sidore C, Kwong A, Timpson N, Koskinen S, Vrieze S, Scott LJ, Zhang H, Mahajan A, Veldink J, Peters U, Pato C, van Duijn CM, Gillies CE, Gandin I, Mezzavilla M, Gilly A, Cocca M, Traglia M, Angius A, Barrett JC, Boomsma D, Branham K, Breen G, Brummett CM, Busonero F, Campbell H, Chan A, Chen S, Chew E, Collins FS, Corbin LJ, Smith GD, Dedoussis G, Dorr M, Farmaki A, Ferrucci L, Forer L, Fraser RM, Gabriel S, Levy S, Groop L, Harrison T, Hattersley A, Holmen OL, Hveem K, Kretzler M, Lee JC, McGue M, Meitinger T, Melzer D, Min JL, Mohlke KL, Vincent JB, Nauck M, Nickerson D, Palotie A, Pato M, Pirastu N, McInnis M, Richards JB, Sala C, Salomaa V, Schlessinger D, Schoenherr S, Slagboom PE, Small K, Spector T, Stambolian D, Tuke M, Tuomilehto J, Van den Berg LH, Van Rheenen W, Volker U, Wijmenga C, Toniolo D, Zeggini E, Gasparini P, Sampson MG, Wilson JF, Frayling T, de Bakker PIW, Swertz MA, McCarroll S, Kooperberg C, Dekker A, Altshuler D, Willer C, Iacono W, Ripatti S, Soranzo N, Walter K, Swaroop A, Cucca F, Anderson CA, Myers RM, Boehnke M, McCarthy MI, Durbin R, Abecasis G, Marchini J, the Haplotype RC (2016) A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet 48:1279–83. https://doi.org/10.1038/ng.3643
Huang J, Howie B, McCarthy S, Memari Y, Walter K, Min JL, Danecek P, Malerba G, Trabetti E, Zheng H, Al Turki S, Amuzu A, Anderson CA, Anney R, Antony D, Artigas MS, Ayub M, Bala S, Barrett JC, Barroso I, Beales P, Benn M, Bentham J, Bhattacharya S, Birney E, Blackwood D, Bobrow M, Bochukova E, Bolton PF, Bounds R, Boustred C, Breen G, Calissano M, Carss K, Pablo Casas J, Chambers JC, Charlton R, Chatterjee K, Chen L, Ciampi A, Cirak S, Clapham P, Clement G, Coates G, Cocca M, Collier DA, Cosgrove C, Cox T, Craddock N, Crooks L, Curran S, Curtis D, Daly A, Day INM, Day-Williams A, Dedoussis G, Down T, Du Y, van Duijn CM, Dunham I, Edkins S, Ekong R, Ellis P, Evans DM, Farooqi IS, Fitzpatrick DR, Flicek P, Floyd J, Foley AR, Franklin CS, Futema M, Gallagher L, Gasparini P, Gaunt TR, Geihs M, Geschwind D, Greenwood C, Griffin H, Grozeva D, Guo X, Guo X, Gurling H, Hart D, Hendricks AE, Holmans P, Huang L, Hubbard T, Humphries SE, Hurles ME, Hysi P, Iotchkova V, Isaacs A, Jackson DK, Jamshidi Y, Johnson J, Joyce C, Karczewski KJ, Kaye J, Keane T, Kemp JP, Kennedy K, Kent A, Keogh J, Khawaja F, Kleber ME, van Kogelenberg M, Kolb-Kokocinski A, Kooner JS, Lachance G, Langenberg C, Langford C, Lawson D, Lee I, van Leeuwen EM, Lek M, Li R, Li Y, Liang J, Lin H, Liu R, Lönnqvist J, Lopes LR, Lopes M, Luan J, MacArthur DG, Mangino M, Marenne G, März W, Maslen J, Matchan A, Mathieson I, McGuffin P, McIntosh AM, McKechanie AG, McQuillin A, Metrustry S, Migone N, Mitchison HM, Moayyeri A, Morris J, Morris R, Muddyman D, Muntoni F, Nordestgaard BG, Northstone K, O’Donovan MC, O’Rahilly S, Onoufriadis A, Oualkacha K, Owen MJ, Palotie A, Panoutsopoulou K, Parker V, Parr JR, Paternoster L, Paunio T, Payne F, Payne SJ, Perry JRB, Pietilainen O, Plagnol V, Pollitt RC, Povey S, Quail MA, Quaye L, Raymond L, Rehnström K, Ridout CK, Ring S, Ritchie GRS, Roberts N, Robinson RL, Savage DB, Scambler P, Schiffels S, Schmidts M, Schoenmakers N, Scott RH, Scott RA, Semple RK, Serra E, Sharp SI, Shaw A, Shihab HA, Shin S, Skuse D, Small KS, Smee C, Smith GD, Southam L, Spasic-Boskovic O, Spector TD, St Clair D, St Pourcain B, Stalker J, Stevens E, Sun J, Surdulescu G, Suvisaari J, Syrris P, Tachmazidou I, Taylor R, Tian J, Tobin MD, Toniolo D, Traglia M, Tybjaerg-Hansen A, Valdes AM, Vandersteen AM, Varbo A, Vijayarangakannan P, Visscher PM, Wain LV, Walters JTR, Wang G, Wang J, Wang Y, Ward K, Wheeler E, Whincup P, Whyte T, Williams HJ, Williamson KA, Wilson C, Wilson SG, Wong K, Xu C, Yang J, Zaza G, Zeggini E, Zhang F, Zhang P, Zhang W, Gambaro G, Richards JB, Durbin R, Timpson NJ, Marchini J, Soranzo N, Consortium U (2015) Improved imputation of low-frequency and rare variants using the UK10K haplotype reference panel. Nat Commun 6:8111. https://doi.org/10.1038/ncomms9111
Lam M, Chen C, Li Z, Martin AR, Bryois J, Ma X, Gaspar H, Ikeda M, Benyamin B, Brown BC, Liu R, Zhou W, Guan L, Kamatani Y, Kim S, Kubo M, Kusumawardhani Agung A. A. A, Liu C, Ma H, Periyasamy S, Takahashi A, Xu Z, Yu H, Zhu F, Chen WJ, Faraone S, Glatt SJ, He L, Hyman SE, Hwu H, McCarroll SA, Neale BM, Sklar P, Wildenauer DB, Yu X, Zhang D, Mowry BJ, Lee J, Holmans P, Xu S, Sullivan PF, Ripke S, O’Donovan MC, Daly MJ, Qin S, Sham P, Iwata N, Hong KS, Schwab SG, Yue W, Tsuang M, Liu J, Ma X, Kahn RS, Shi Y, Huang H, Schizophrenia Working Group of the Psychiatric, Genomics Consortium, Indonesia SC, Genetic REsearch on schizophreniA neTwork-China and the Netherlands, (GREAT-CN) (2019) Comparative genetic architectures of schizophrenia in East Asian and European populations. Nat Genet 51:1670–8. https://doi.org/10.1038/s41588-019-0512-x
Privé F, Arbel J, Vilhjálmsson BJ (2020) LDpred2: better, faster, stronger. Bioinformatics 36:5424–5431. https://doi.org/10.1093/bioinformatics/btaa1029
Kim MS, Patel KP, Teng AK, Berens AJ, Lachance J (2018) Genetic disease risks can be misestimated across global populations. Genome Biol 19:179. https://doi.org/10.1186/s13059-018-1561-7
Acknowledgements
The study was supported by the National Institute of General Medical Sciences under award number P20GM121325 and by the National Institute on Minority Health and Health Disparities of the National Institutes of Health under award number R21MD013681. In addition, the National Supercomputing Institute at the University of Nevada, Las Vegas, provided facilities for bioinformatical analysis in this study. Part of Dr. Qing Wu’s work was completed at the Nevada Institute of Personalized Medicine, College of Sciences, Department of Epidemiology and Biostatistics, School of Public Health, University of Nevada, Las Vegas.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethics approval
This research work was approved by the UK Biobank and the institutional review board at the University of Nevada, Las Vegas. This study has been conducted using the UK Biobank Data Resource under application number 58122.
Conflicts of interest
None.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Xiao, X., Wu, Q. Ethnic disparities in fracture risk assessment using polygenic scores. Osteoporos Int 34, 943–953 (2023). https://doi.org/10.1007/s00198-023-06712-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00198-023-06712-y